nimare.decode.continuous¶
Methods for decoding unthresholded brain maps into text.
Functions
corr_decode(dataset, img[, features, …]) |
|
||
corr_dist_decode(dataset, img[, features, …]) |
Builds feature-specific distributions of correlations with input image for image-based meta-analytic functional decoding. | ||
gclda_decode_map(model, image[, …]) |
Perform image-to-text decoding for continuous inputs (e.g., unthresholded statistical maps), according to the method described in [1]. |
-
corr_decode(dataset, img, features=None, frequency_threshold=0.001, meta_estimator=None, target_image='specificity_z')[source]¶ Parameters: - dataset (
nimare.dataset.Dataset) – A dataset with coordinates. - img (
nibabel.Nifti1.Nifti1Image) – Input image to decode. Must have same affine/dimensions as dataset mask. - features (
list, optional) – List of features in dataset annotations to use for decoding. Default is None, which uses all features available. - frequency_threshold (
float, optional) – Threshold to apply to dataset annotations. Values greater than or equal to the threshold as assigned as label+, while values below the threshold are considered label-. Default is 0.001. - meta_estimator (initialized
nimare.meta.cbma.base.CBMAEstimator, optional) – Defaults to MKDAChi2. - target_image (
str, optional) – Image frommeta_estimator’s results to use for decoding. Dependent on estimator.
Returns: out_df – A DataFrame with two columns: ‘feature’ (label) and ‘r’ (correlation coefficient). There will be one row for each feature.
Return type: - dataset (
-
corr_dist_decode(dataset, img, features=None, frequency_threshold=0.001, target_image='z')[source]¶ Builds feature-specific distributions of correlations with input image for image-based meta-analytic functional decoding.
Parameters: - dataset (
nimare.dataset.Dataset) – A dataset with images. - img (
nibabel.Nifti1.Nifti1Image) – Input image to decode. Must have same affine/dimensions as dataset mask. - features (
list, optional) – List of features in dataset annotations to use for decoding. Default is None, which uses all features available. - frequency_threshold (
float, optional) – Threshold to apply to dataset annotations. Values greater than or equal to the threshold as assigned as label+, while values below the threshold are considered label-. Default is 0.001. - target_image ({'z', 'con'}, optional) – Image type from database to use for decoding.
Returns: out_df – DataFrame with a row for each feature used for decoding and two columns: mean and std. Values describe the distributions of correlation coefficients (in terms of Fisher-transformed z-values).
Return type: - dataset (
-
gclda_decode_map(model, image, topic_priors=None, prior_weight=1)[source]¶ Perform image-to-text decoding for continuous inputs (e.g., unthresholded statistical maps), according to the method described in [1].
Parameters: - model (
nimare.annotate.topic.GCLDAModel) – Model object needed for decoding. - image (
nibabel.nifti1.Nifti1Imageorstr) – Whole-brain image to decode into text. Must be in same space as model and dataset. Model’s template available in model.dataset.mask_img. - topic_priors (
numpy.ndarrayoffloat, optional) – A 1d array of size (n_topics) with values for topic weighting. If None, no weighting is done. Default is None. - prior_weight (
float, optional) – The weight by which the prior will affect the decoding. Default is 1.
Returns: - decoded_df (
pandas.DataFrame) – A DataFrame with the word-tokens and their associated weights. - topic_weights (
numpy.ndarrayoffloat) – The weights of the topics used in decoding.
Notes
Notation Meaning 
Voxel 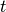
Topic 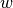
Word type 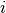
Input image 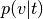
Probability of topic given voxel ( p_topic_g_voxel)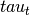
Topic weight vector ( topic_weights)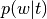
Probability of word type given topic ( p_word_g_topic)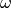
1d array from input image ( input_values)Compute
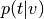 (
(p_topic_g_voxel).- From
gclda.model.Model.get_spatial_probs()
- From
Squeeze input image to 1d array
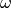 (
(input_values).Compute topic weight vector (
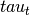 ) by multiplying
) by multiplying
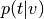 by input image.
by input image.Multiply
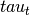 by
by
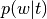 .
.The resulting vector (
word_weights) reflects arbitrarily scaled term weights for the input image.
References
[1] (1, 2) Rubin, Timothy N., et al. “Decoding brain activity using a large-scale probabilistic functional-anatomical atlas of human cognition.” PLoS computational biology 13.10 (2017): e1005649. https://doi.org/10.1371/journal.pcbi.1005649 - model (