Note
Click here to download the full example code
Run image-based meta-analyses on 21 pain studies¶
Collection of NIDM-Results packs downloaded from Neurovault collection 1425, uploaded by Dr. Camille Maumet.
Caution
Dataset querying will likely change as we work to shift database querying to a remote database, rather than handling it locally with NiMARE.
import os
import numpy as np
import nibabel as nib
from nilearn.masking import apply_mask, unmask
from nilearn.plotting import plot_stat_map
import nimare
from nimare.tests.utils import get_test_data_path
from nimare.meta.esma import fishers
from nimare.meta.ibma import (Fishers, Stouffers, WeightedStouffers, RFX_GLM)
Download data¶
Load Dataset¶
dset_file = os.path.join(get_test_data_path(), 'nidm_pain_dset.json')
dset = nimare.dataset.Dataset(dset_file)
dset.update_path(dset_dir)
logp_thresh = -np.log(.05)
Fisher’s (using functions)¶
Get images for analysis
files = dset.get_images(imtype='z')
files = [f for f in files if f]
z_data = dset.masker.transform(files)
print('{0} studies found.'.format(z_data.shape[0]))
result = fishers(z_data)
fishers_result = dset.masker.inverse_transform(result['z'])
plot_stat_map(fishers_result, cut_coords=[0, 0, -8],
draw_cross=False, cmap='RdBu_r')
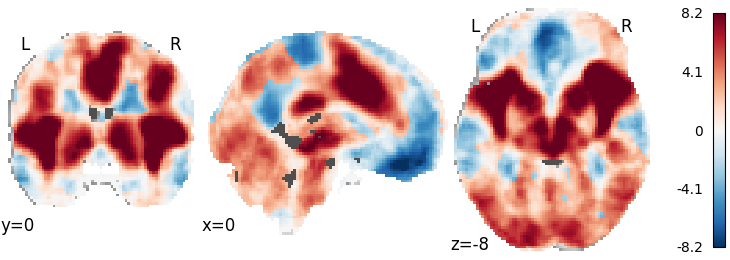
Out:
11 studies found.
<nilearn.plotting.displays.OrthoSlicer object at 0x7f0f799f4048>
Fisher’s (using Estimators)¶
Here is the object-oriented approach
meta = Fishers()
meta.fit(dset)
plot_stat_map(meta.results.get_map('z'), cut_coords=[0, 0, -8],
draw_cross=False, cmap='RdBu_r')
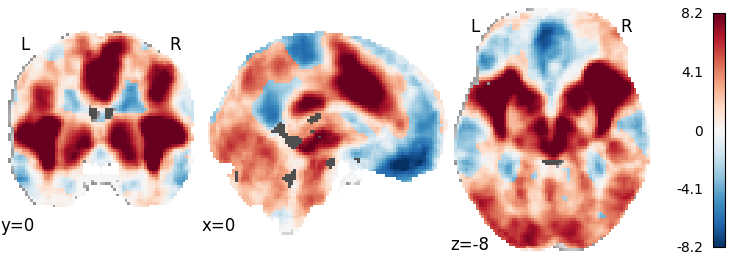
Out:
<nilearn.plotting.displays.OrthoSlicer object at 0x7f0f7906bd30>
Stouffer’s with fixed-effects inference¶
meta = Stouffers(inference='ffx', null='theoretical', n_iters=None)
meta.fit(dset)
plot_stat_map(meta.results.get_map('z'), cut_coords=[0, 0, -8],
draw_cross=False, cmap='RdBu_r')
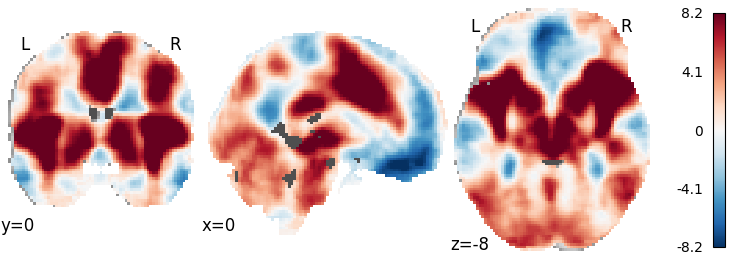
Out:
<nilearn.plotting.displays.OrthoSlicer object at 0x7f0f79d0a588>
Stouffer’s with random-effects inference using theoretical null distribution¶
meta = Stouffers(inference='rfx', null='theoretical', n_iters=None)
meta.fit(dset)
plot_stat_map(meta.results.get_map('z'), cut_coords=[0, 0, -8],
draw_cross=False, cmap='RdBu_r')
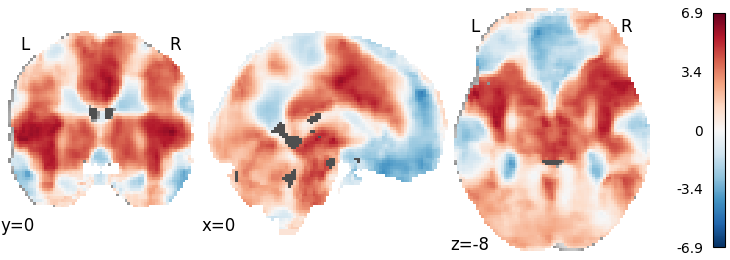
Out:
/home/docs/.pyenv/versions/3.7.3/lib/python3.7/site-packages/scipy/stats/_distn_infrastructure.py:903: RuntimeWarning: invalid value encountered in greater
return (a < x) & (x < b)
/home/docs/.pyenv/versions/3.7.3/lib/python3.7/site-packages/scipy/stats/_distn_infrastructure.py:903: RuntimeWarning: invalid value encountered in less
return (a < x) & (x < b)
/home/docs/.pyenv/versions/3.7.3/lib/python3.7/site-packages/scipy/stats/_distn_infrastructure.py:1912: RuntimeWarning: invalid value encountered in less_equal
cond2 = cond0 & (x <= _a)
<nilearn.plotting.displays.OrthoSlicer object at 0x7f0f7999afd0>
Stouffer’s with random-effects inference using empirical null distribution¶
meta = Stouffers(inference='rfx', null='empirical', n_iters=100)
meta.fit(dset)
plot_stat_map(meta.results.get_map('z'), cut_coords=[0, 0, -8],
draw_cross=False, cmap='RdBu_r')
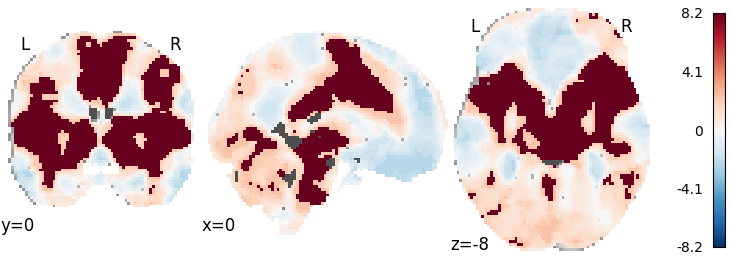
Out:
<nilearn.plotting.displays.OrthoSlicer object at 0x7f0f79f41358>
Weighted Stouffer’s¶
meta = WeightedStouffers()
meta.fit(dset)
plot_stat_map(meta.results.get_map('z'), cut_coords=[0, 0, -8],
draw_cross=False, cmap='RdBu_r')
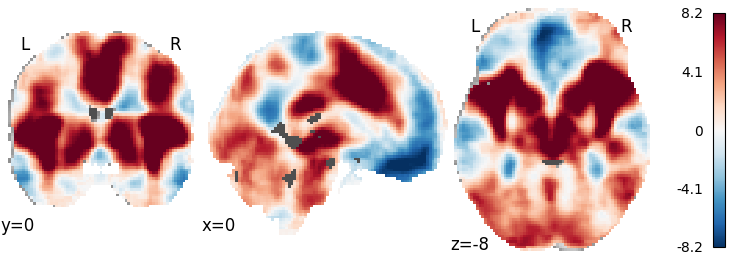
Out:
<nilearn.plotting.displays.OrthoSlicer object at 0x7f0f79dd06d8>
RFX GLM with theoretical null distribution¶
meta = RFX_GLM(null='theoretical', n_iters=None)
meta.fit(dset)
plot_stat_map(meta.results.get_map('z'), cut_coords=[0, 0, -8],
draw_cross=False, cmap='RdBu_r')
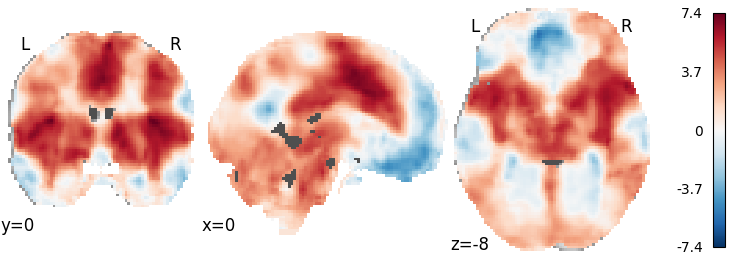
Out:
<nilearn.plotting.displays.OrthoSlicer object at 0x7f0f79ddc080>
RFX GLM with empirical null distribution¶
meta = RFX_GLM(null='empirical', n_iters=100)
meta.fit(dset)
plot_stat_map(meta.results.get_map('z'), cut_coords=[0, 0, -8],
draw_cross=False, cmap='RdBu_r')
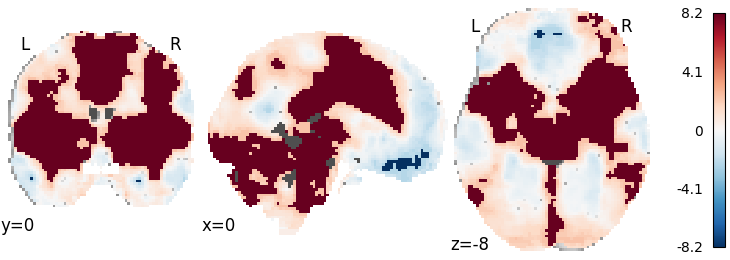
Out:
<nilearn.plotting.displays.OrthoSlicer object at 0x7f0f799a4c50>
Total running time of the script: ( 0 minutes 49.334 seconds)