Note
Click here to download the full example code
Generate modeled activation maps
For coordinate-based data, individual studies’ statistical maps are mimicked by generating “modeled activation” (MA) maps. These MA maps are used in the CBMA algorithms, although the specific method used to generate the MA maps differs by algorithm.
Start with the necessary imports
import os
import matplotlib.pyplot as plt
import numpy as np
from nilearn.plotting import plot_stat_map
import nimare
from nimare.tests.utils import get_test_data_path
Load Dataset
dset_file = os.path.join(get_test_data_path(), "nidm_pain_dset.json")
dset = nimare.dataset.Dataset(dset_file)
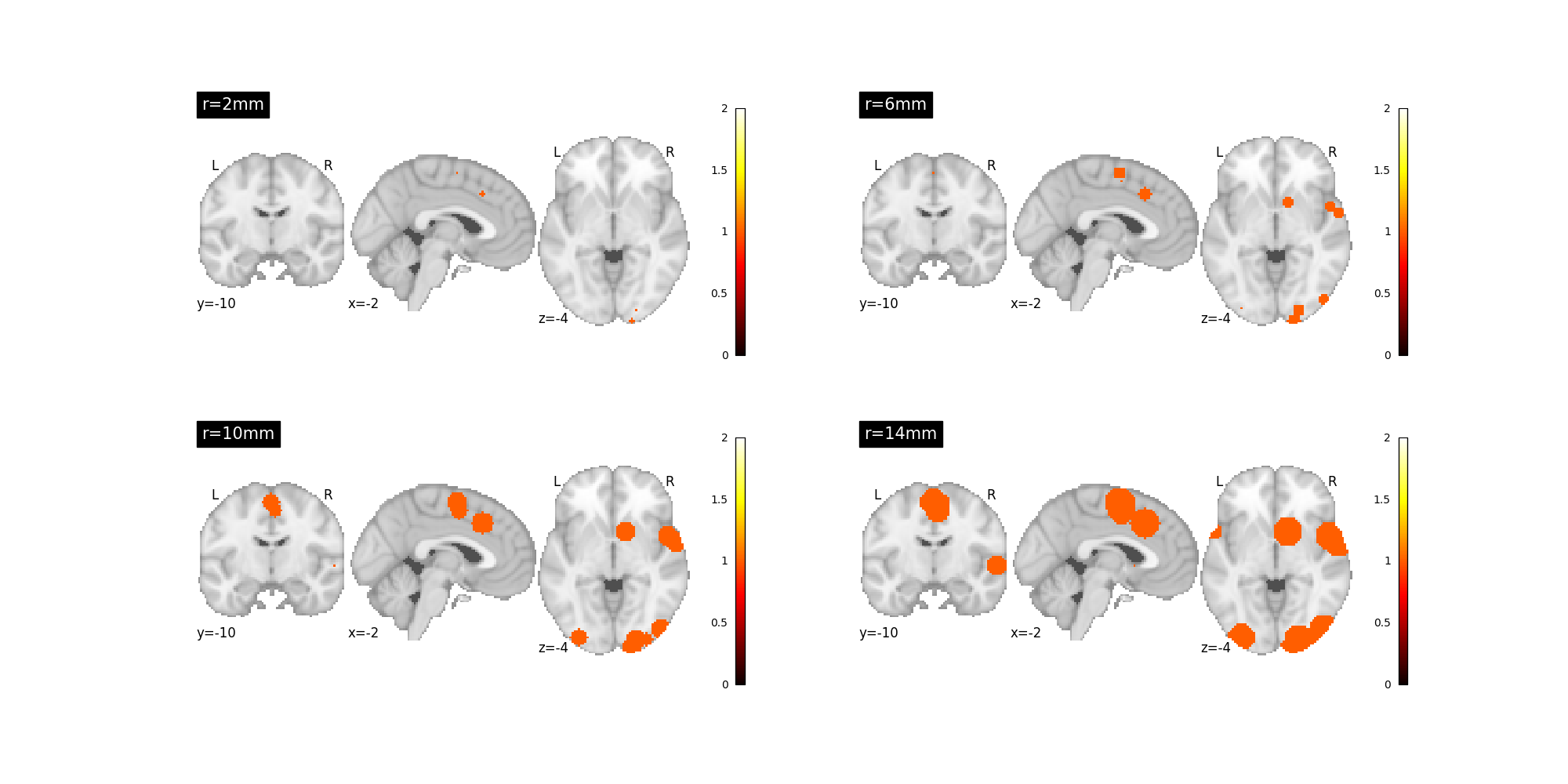
Each kernel can taken certain parameters that control behavior
For example, nimare.meta.kernel.MKDAKernel kernel accepts an r
argument to control the radius of the kernel.
kernel = nimare.meta.kernel.MKDAKernel(r=2)
mkda_r02 = kernel.transform(dset, return_type="image")
kernel = nimare.meta.kernel.MKDAKernel(r=6)
mkda_r06 = kernel.transform(dset, return_type="image")
kernel = nimare.meta.kernel.MKDAKernel(r=10)
mkda_r10 = kernel.transform(dset, return_type="image")
kernel = nimare.meta.kernel.MKDAKernel(r=14)
mkda_r14 = kernel.transform(dset, return_type="image")
fig, axes = plt.subplots(nrows=2, ncols=2, figsize=(20, 10))
plot_stat_map(
mkda_r02[2], cut_coords=[-2, -10, -4], title="r=2mm", vmax=2, axes=axes[0, 0], draw_cross=False
)
plot_stat_map(
mkda_r06[2], cut_coords=[-2, -10, -4], title="r=6mm", vmax=2, axes=axes[0, 1], draw_cross=False
)
plot_stat_map(
mkda_r10[2],
cut_coords=[-2, -10, -4],
title="r=10mm",
vmax=2,
axes=axes[1, 0],
draw_cross=False,
)
plot_stat_map(
mkda_r14[2],
cut_coords=[-2, -10, -4],
title="r=14mm",
vmax=2,
axes=axes[1, 1],
draw_cross=False,
)
fig.show()
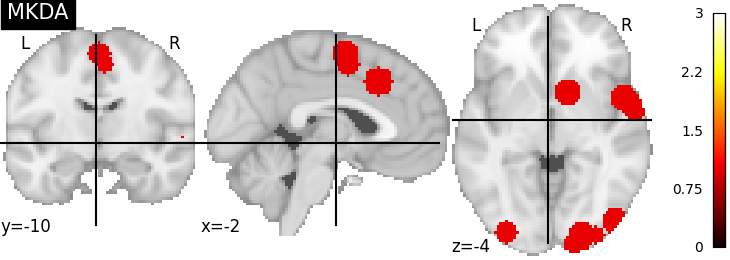
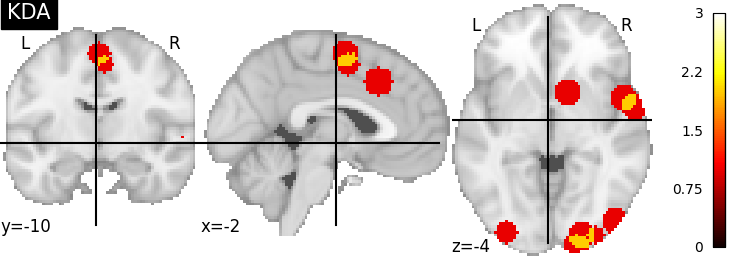
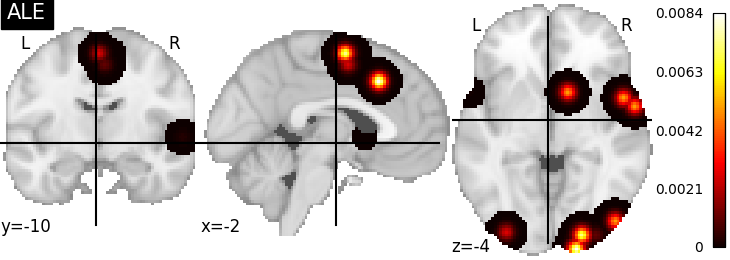
There are several kernels available
nimare.meta.kernel.MKDAKernel convolves coordinates with a
sphere and takes the union across voxels.
nimare.meta.kernel.KDAKernel convolves coordinates with a
sphere as well, but takes the sum across voxels.
nimare.meta.kernel.ALEKernel convolves coordinates with a 3D
Gaussian, for which the FWHM is determined by the sample size of each study.
kernel = nimare.meta.kernel.MKDAKernel(r=10)
mkda_res = kernel.transform(dset, return_type="image")
kernel = nimare.meta.kernel.KDAKernel(r=10)
kda_res = kernel.transform(dset, return_type="image")
kernel = nimare.meta.kernel.ALEKernel(sample_size=20)
ale_res = kernel.transform(dset, return_type="image")
max_conv = np.max(kda_res[2].get_fdata())
plot_stat_map(mkda_res[2], cut_coords=[-2, -10, -4], title="MKDA", vmax=max_conv)
plot_stat_map(kda_res[2], cut_coords=[-2, -10, -4], title="KDA", vmax=max_conv)
plot_stat_map(ale_res[2], cut_coords=[-2, -10, -4], title="ALE")
Out:
<nilearn.plotting.displays.OrthoSlicer object at 0x7f1889b3b4d0>
Total running time of the script: ( 0 minutes 11.448 seconds)