Note
Click here to download the full example code
Run coordinate-based meta-analyses on 21 pain studies
Collection of NIDM-Results packs downloaded from Neurovault collection 1425, uploaded by Dr. Camille Maumet.
Note
Creation of the Dataset from the NIDM-Results packs was done with custom code. The Results packs for collection 1425 are not completely NIDM-Results-compliant, so the nidmresults library could not be used to facilitate data extraction.
import os
from nilearn.plotting import plot_stat_map
import nimare
from nimare.tests.utils import get_test_data_path
Load Dataset
dset_file = os.path.join(get_test_data_path(), "nidm_pain_dset.json")
dset = nimare.dataset.Dataset(dset_file)
mask_img = dset.masker.mask_img
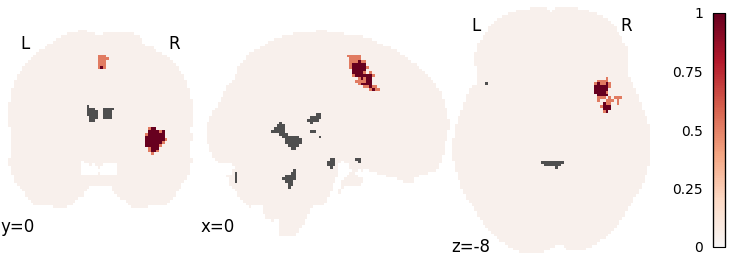
MKDA density analysis
mkda = nimare.meta.MKDADensity(kernel__r=10, null_method="approximate")
mkda.fit(dset)
corr = nimare.correct.FWECorrector(method="montecarlo", n_iters=10, n_cores=1)
cres = corr.transform(mkda.results)
plot_stat_map(
cres.get_map("logp_level-voxel_corr-FWE_method-montecarlo"),
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
)
Out:
0%| | 0/10 [00:00<?, ?it/s]
10%|# | 1/10 [00:00<00:02, 3.47it/s]
20%|## | 2/10 [00:00<00:02, 3.48it/s]
30%|### | 3/10 [00:00<00:02, 3.49it/s]
40%|#### | 4/10 [00:01<00:01, 3.50it/s]
50%|##### | 5/10 [00:01<00:01, 3.50it/s]
60%|###### | 6/10 [00:01<00:01, 3.49it/s]
70%|####### | 7/10 [00:02<00:00, 3.50it/s]
80%|######## | 8/10 [00:02<00:00, 3.51it/s]
90%|######### | 9/10 [00:02<00:00, 3.51it/s]
100%|##########| 10/10 [00:02<00:00, 3.51it/s]
100%|##########| 10/10 [00:02<00:00, 3.50it/s]
<nilearn.plotting.displays.OrthoSlicer object at 0x7fe67941e810>
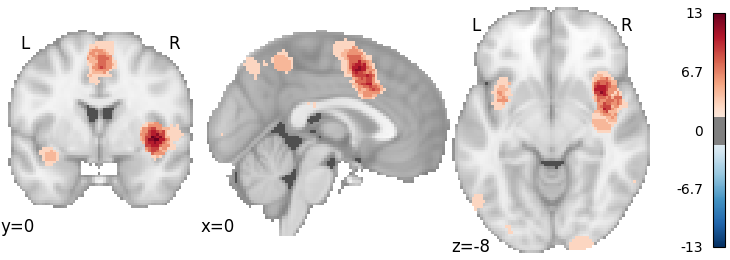
MKDA Chi2 with FDR correction
mkda = nimare.meta.MKDAChi2(kernel__r=10)
dset1 = dset.slice(dset.ids)
dset2 = dset.slice(dset.ids)
mkda.fit(dset1, dset2)
corr = nimare.correct.FDRCorrector(method="bh", alpha=0.001)
cres = corr.transform(mkda.results)
plot_stat_map(
cres.get_map("z_desc-consistency_level-voxel_corr-FDR_method-bh"),
threshold=1.65,
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
)
Out:
/home/docs/checkouts/readthedocs.org/user_builds/nimare/checkouts/0.0.11rc1/nimare/meta/cbma/mkda.py:259: RuntimeWarning: invalid value encountered in true_divide
pFgA = pAgF * pF / pA
/home/docs/checkouts/readthedocs.org/user_builds/nimare/checkouts/0.0.11rc1/nimare/meta/cbma/mkda.py:263: RuntimeWarning: invalid value encountered in true_divide
pFgA_prior = pAgF * self.prior / pAgF_prior
<nilearn.plotting.displays.OrthoSlicer object at 0x7fe678d09510>
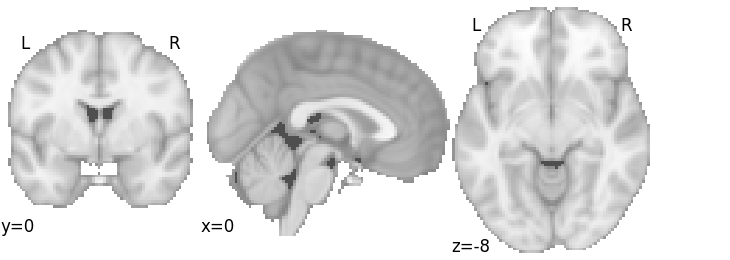
MKDA Chi2 with FWE correction
Since we’ve already fitted the Estimator, we can just apply a new Corrector to the estimator.
corr = nimare.correct.FWECorrector(method="montecarlo", n_iters=10, n_cores=1)
cres = corr.transform(mkda.results)
plot_stat_map(
cres.get_map("z_desc-consistency_level-voxel_corr-FWE_method-montecarlo"),
threshold=1.65,
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
)
Out:
0%| | 0/10 [00:00<?, ?it/s]
10%|# | 1/10 [00:00<00:03, 2.85it/s]
20%|## | 2/10 [00:00<00:02, 2.85it/s]
30%|### | 3/10 [00:01<00:02, 2.86it/s]
40%|#### | 4/10 [00:01<00:02, 2.86it/s]
50%|##### | 5/10 [00:01<00:01, 2.87it/s]
60%|###### | 6/10 [00:02<00:01, 2.87it/s]
70%|####### | 7/10 [00:02<00:01, 2.87it/s]
80%|######## | 8/10 [00:02<00:00, 2.88it/s]
90%|######### | 9/10 [00:03<00:00, 2.88it/s]
100%|##########| 10/10 [00:03<00:00, 2.88it/s]
100%|##########| 10/10 [00:03<00:00, 2.87it/s]
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.11rc1/lib/python3.7/site-packages/nilearn/plotting/displays.py:880: UserWarning: empty mask
get_mask_bounds(new_img_like(img, not_mask, affine))
<nilearn.plotting.displays.OrthoSlicer object at 0x7fe679660450>
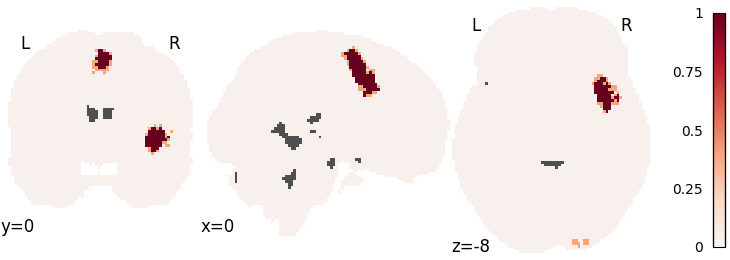
KDA
kda = nimare.meta.KDA(kernel__r=10, null_method="approximate")
kda.fit(dset)
corr = nimare.correct.FWECorrector(method="montecarlo", n_iters=10, n_cores=1)
cres = corr.transform(kda.results)
plot_stat_map(
cres.get_map("logp_level-voxel_corr-FWE_method-montecarlo"),
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
)
Out:
0%| | 0/10 [00:00<?, ?it/s]
10%|# | 1/10 [00:00<00:02, 3.25it/s]
20%|## | 2/10 [00:00<00:02, 3.32it/s]
30%|### | 3/10 [00:00<00:02, 3.34it/s]
40%|#### | 4/10 [00:01<00:01, 3.35it/s]
50%|##### | 5/10 [00:01<00:01, 3.36it/s]
60%|###### | 6/10 [00:01<00:01, 3.35it/s]
70%|####### | 7/10 [00:02<00:00, 3.36it/s]
80%|######## | 8/10 [00:02<00:00, 3.35it/s]
90%|######### | 9/10 [00:02<00:00, 3.35it/s]
100%|##########| 10/10 [00:02<00:00, 3.35it/s]
100%|##########| 10/10 [00:02<00:00, 3.35it/s]
<nilearn.plotting.displays.OrthoSlicer object at 0x7fe679416ed0>
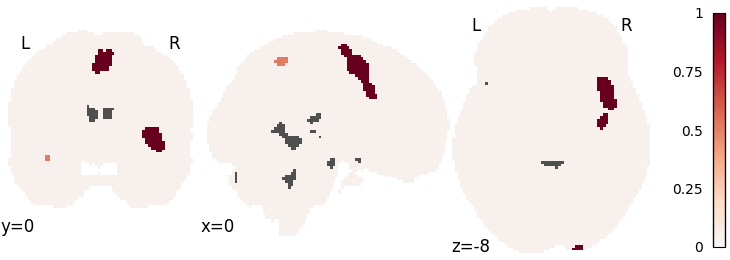
ALE
ale = nimare.meta.ALE(null_method="approximate")
ale.fit(dset)
corr = nimare.correct.FWECorrector(method="montecarlo", n_iters=10, n_cores=1)
cres = corr.transform(ale.results)
plot_stat_map(
cres.get_map("logp_level-cluster_corr-FWE_method-montecarlo"),
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
)
Out:
0%| | 0/10 [00:00<?, ?it/s]
10%|# | 1/10 [00:00<00:03, 2.77it/s]
20%|## | 2/10 [00:00<00:02, 2.79it/s]
30%|### | 3/10 [00:01<00:02, 2.80it/s]
40%|#### | 4/10 [00:01<00:02, 2.80it/s]
50%|##### | 5/10 [00:01<00:01, 2.81it/s]
60%|###### | 6/10 [00:02<00:01, 2.80it/s]
70%|####### | 7/10 [00:02<00:01, 2.82it/s]
80%|######## | 8/10 [00:02<00:00, 2.83it/s]
90%|######### | 9/10 [00:03<00:00, 2.83it/s]
100%|##########| 10/10 [00:03<00:00, 2.84it/s]
100%|##########| 10/10 [00:03<00:00, 2.82it/s]
<nilearn.plotting.displays.OrthoSlicer object at 0x7fe679621310>
Total running time of the script: ( 0 minutes 34.926 seconds)