Note
Click here to download the full example code
Combine CBMA kernels and estimators
Collection of NIDM-Results packs downloaded from Neurovault collection 1425, uploaded by Dr. Camille Maumet.
Note
Creation of the Dataset from the NIDM-Results packs was done with custom code. The Results packs for collection 1425 are not completely NIDM-Results-compliant, so the nidmresults library could not be used to facilitate data extraction.
import os
from nilearn.plotting import plot_stat_map
from nimare.correct import FWECorrector
from nimare.dataset import Dataset
from nimare.meta import ALE, KDA, MKDAChi2, MKDADensity
from nimare.meta.kernel import ALEKernel, KDAKernel, MKDAKernel
from nimare.utils import get_resource_path
Load Dataset
dset_file = os.path.join(get_resource_path(), "nidm_pain_dset.json")
dset = Dataset(dset_file)
mask_img = dset.masker.mask_img
List possible kernel transformers
kernel_transformers = {
"MKDA kernel": MKDAKernel,
"KDA kernel": KDAKernel,
"ALE kernel": ALEKernel,
}
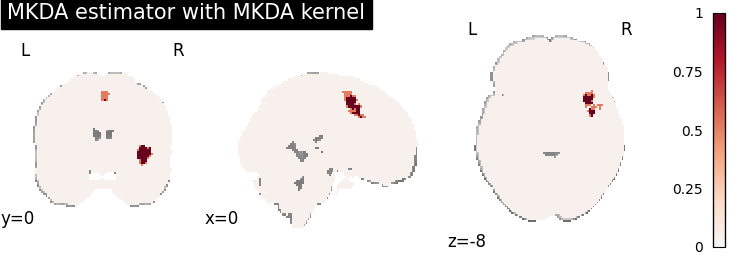
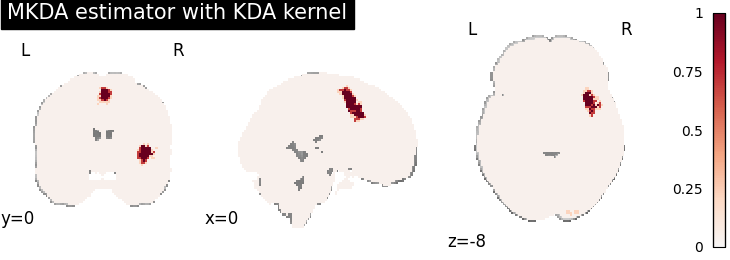
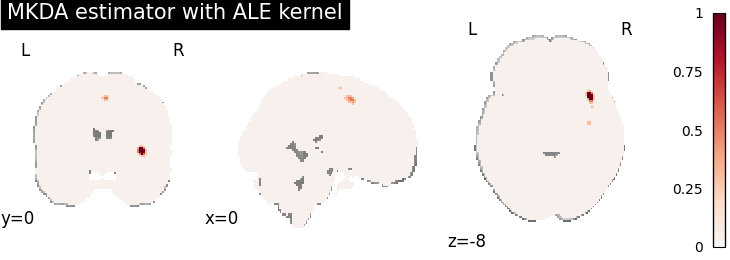
MKDA density analysis
for kt_name, kt in kernel_transformers.items():
try:
mkda = MKDADensity(kernel_transformer=kt, null_method="approximate")
mkda.fit(dset)
corr = FWECorrector(method="montecarlo", n_iters=10, n_cores=1)
cres = corr.transform(mkda.results)
plot_stat_map(
cres.get_map("logp_level-voxel_corr-FWE_method-montecarlo"),
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
title="MKDA estimator with %s" % kt_name,
)
except AttributeError:
print(
"\nError: the %s does not currently work with the MKDA meta-analysis method\n"
% kt_name
)
Out:
0%| | 0/10 [00:00<?, ?it/s]
10%|# | 1/10 [00:00<00:02, 3.42it/s]
20%|## | 2/10 [00:00<00:02, 3.45it/s]
30%|### | 3/10 [00:00<00:02, 3.43it/s]
40%|#### | 4/10 [00:01<00:01, 3.44it/s]
50%|##### | 5/10 [00:01<00:01, 3.42it/s]
60%|###### | 6/10 [00:01<00:01, 3.39it/s]
70%|####### | 7/10 [00:02<00:00, 3.39it/s]
80%|######## | 8/10 [00:02<00:00, 3.41it/s]
90%|######### | 9/10 [00:02<00:00, 3.41it/s]
100%|##########| 10/10 [00:02<00:00, 3.41it/s]
100%|##########| 10/10 [00:02<00:00, 3.41it/s]
0%| | 0/10 [00:00<?, ?it/s]
10%|# | 1/10 [00:00<00:02, 3.19it/s]
20%|## | 2/10 [00:00<00:02, 3.19it/s]
30%|### | 3/10 [00:00<00:02, 3.15it/s]
40%|#### | 4/10 [00:01<00:01, 3.15it/s]
50%|##### | 5/10 [00:01<00:01, 3.16it/s]
60%|###### | 6/10 [00:01<00:01, 3.15it/s]
70%|####### | 7/10 [00:02<00:00, 3.17it/s]
80%|######## | 8/10 [00:02<00:00, 3.18it/s]
90%|######### | 9/10 [00:02<00:00, 3.18it/s]
100%|##########| 10/10 [00:03<00:00, 3.18it/s]
100%|##########| 10/10 [00:03<00:00, 3.17it/s]
0%| | 0/10 [00:00<?, ?it/s]
10%|# | 1/10 [00:00<00:02, 3.17it/s]
20%|## | 2/10 [00:00<00:02, 3.20it/s]
30%|### | 3/10 [00:00<00:02, 3.20it/s]
40%|#### | 4/10 [00:01<00:01, 3.20it/s]
50%|##### | 5/10 [00:01<00:01, 3.19it/s]
60%|###### | 6/10 [00:01<00:01, 3.19it/s]
70%|####### | 7/10 [00:02<00:00, 3.19it/s]
80%|######## | 8/10 [00:02<00:00, 3.18it/s]
90%|######### | 9/10 [00:02<00:00, 3.19it/s]
100%|##########| 10/10 [00:03<00:00, 3.20it/s]
100%|##########| 10/10 [00:03<00:00, 3.19it/s]
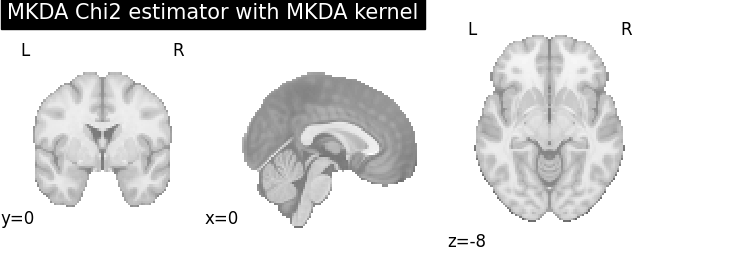
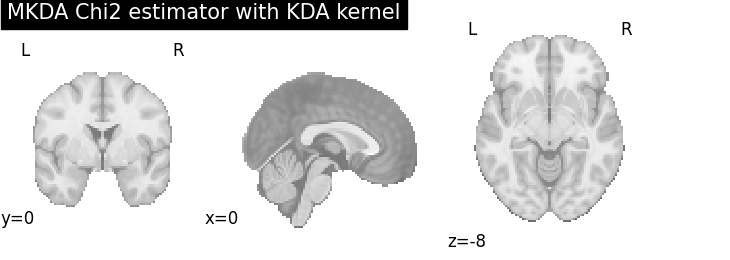
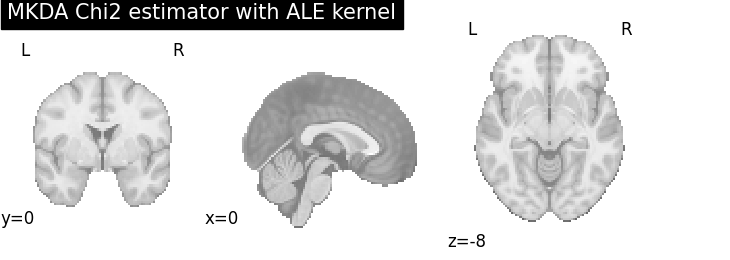
MKDA Chi2
for kt_name, kt in kernel_transformers.items():
try:
mkda = MKDAChi2(kernel_transformer=kt)
dset1 = dset.slice(dset.ids)
dset2 = dset.slice(dset.ids)
mkda.fit(dset1, dset2)
corr = FWECorrector(method="montecarlo", n_iters=10, n_cores=1)
cres = corr.transform(mkda.results)
plot_stat_map(
cres.get_map("z_desc-consistency_level-voxel_corr-FWE_method-montecarlo"),
threshold=1.65,
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
title="MKDA Chi2 estimator with %s" % kt_name,
)
except AttributeError:
print(
"\nError: the %s does not currently work with the MKDA Chi2 meta-analysis method\n"
% kt_name
)
Out:
0%| | 0/10 [00:00<?, ?it/s]
10%|# | 1/10 [00:00<00:02, 3.01it/s]
20%|## | 2/10 [00:00<00:02, 3.00it/s]
30%|### | 3/10 [00:00<00:02, 3.01it/s]
40%|#### | 4/10 [00:01<00:01, 3.01it/s]
50%|##### | 5/10 [00:01<00:01, 3.01it/s]
60%|###### | 6/10 [00:01<00:01, 3.00it/s]
70%|####### | 7/10 [00:02<00:00, 3.01it/s]
80%|######## | 8/10 [00:02<00:00, 3.00it/s]
90%|######### | 9/10 [00:02<00:00, 3.00it/s]
100%|##########| 10/10 [00:03<00:00, 3.01it/s]
100%|##########| 10/10 [00:03<00:00, 3.01it/s]
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.12rc1/lib/python3.7/site-packages/nilearn/plotting/displays/_slicers.py:373: UserWarning: empty mask
get_mask_bounds(new_img_like(img, not_mask, affine))
0%| | 0/10 [00:00<?, ?it/s]
10%|# | 1/10 [00:00<00:03, 2.65it/s]
20%|## | 2/10 [00:00<00:03, 2.66it/s]
30%|### | 3/10 [00:01<00:02, 2.66it/s]
40%|#### | 4/10 [00:01<00:02, 2.66it/s]
50%|##### | 5/10 [00:01<00:01, 2.66it/s]
60%|###### | 6/10 [00:02<00:01, 2.67it/s]
70%|####### | 7/10 [00:02<00:01, 2.67it/s]
80%|######## | 8/10 [00:03<00:00, 2.67it/s]
90%|######### | 9/10 [00:03<00:00, 2.67it/s]
100%|##########| 10/10 [00:03<00:00, 2.67it/s]
100%|##########| 10/10 [00:03<00:00, 2.67it/s]
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.12rc1/lib/python3.7/site-packages/nilearn/plotting/displays/_slicers.py:373: UserWarning: empty mask
get_mask_bounds(new_img_like(img, not_mask, affine))
0%| | 0/10 [00:00<?, ?it/s]
10%|# | 1/10 [00:00<00:03, 2.65it/s]
20%|## | 2/10 [00:00<00:03, 2.65it/s]
30%|### | 3/10 [00:01<00:02, 2.65it/s]
40%|#### | 4/10 [00:01<00:02, 2.66it/s]
50%|##### | 5/10 [00:01<00:01, 2.65it/s]
60%|###### | 6/10 [00:02<00:01, 2.66it/s]
70%|####### | 7/10 [00:02<00:01, 2.66it/s]
80%|######## | 8/10 [00:03<00:00, 2.66it/s]
90%|######### | 9/10 [00:03<00:00, 2.66it/s]
100%|##########| 10/10 [00:03<00:00, 2.67it/s]
100%|##########| 10/10 [00:03<00:00, 2.66it/s]
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.12rc1/lib/python3.7/site-packages/nilearn/plotting/displays/_slicers.py:373: UserWarning: empty mask
get_mask_bounds(new_img_like(img, not_mask, affine))
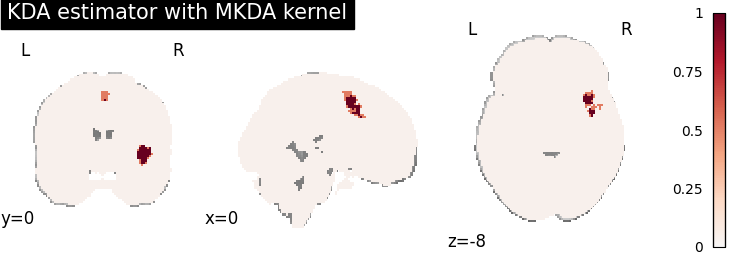
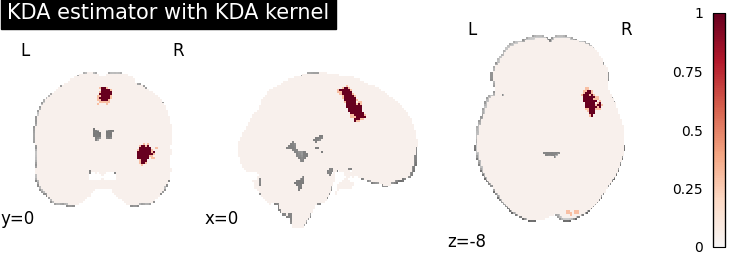
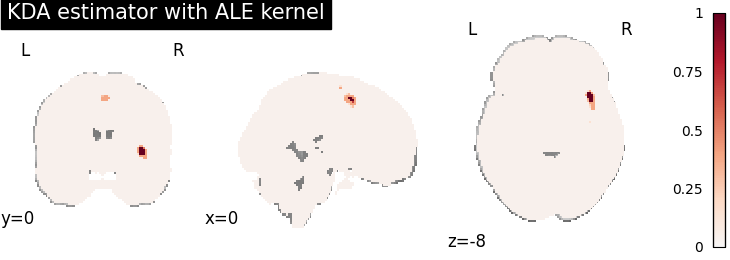
KDA
for kt_name, kt in kernel_transformers.items():
try:
kda = KDA(kernel_transformer=kt, null_method="approximate")
kda.fit(dset)
corr = FWECorrector(method="montecarlo", n_iters=10, n_cores=1)
cres = corr.transform(kda.results)
plot_stat_map(
cres.get_map("logp_level-voxel_corr-FWE_method-montecarlo"),
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
title="KDA estimator with %s" % kt_name,
)
except AttributeError:
print(
"\nError: the %s does not currently work with the KDA meta-analysis method\n" % kt_name
)
Out:
0%| | 0/10 [00:00<?, ?it/s]
10%|# | 1/10 [00:00<00:02, 3.45it/s]
20%|## | 2/10 [00:00<00:02, 3.53it/s]
30%|### | 3/10 [00:00<00:01, 3.55it/s]
40%|#### | 4/10 [00:01<00:01, 3.58it/s]
50%|##### | 5/10 [00:01<00:01, 3.59it/s]
60%|###### | 6/10 [00:01<00:01, 3.61it/s]
70%|####### | 7/10 [00:01<00:00, 3.62it/s]
80%|######## | 8/10 [00:02<00:00, 3.63it/s]
90%|######### | 9/10 [00:02<00:00, 3.63it/s]
100%|##########| 10/10 [00:02<00:00, 3.60it/s]
100%|##########| 10/10 [00:02<00:00, 3.59it/s]
0%| | 0/10 [00:00<?, ?it/s]
10%|# | 1/10 [00:00<00:02, 3.35it/s]
20%|## | 2/10 [00:00<00:02, 3.36it/s]
30%|### | 3/10 [00:00<00:02, 3.37it/s]
40%|#### | 4/10 [00:01<00:01, 3.36it/s]
50%|##### | 5/10 [00:01<00:01, 3.36it/s]
60%|###### | 6/10 [00:01<00:01, 3.35it/s]
70%|####### | 7/10 [00:02<00:00, 3.35it/s]
80%|######## | 8/10 [00:02<00:00, 3.37it/s]
90%|######### | 9/10 [00:02<00:00, 3.39it/s]
100%|##########| 10/10 [00:02<00:00, 3.39it/s]
100%|##########| 10/10 [00:02<00:00, 3.37it/s]
0%| | 0/10 [00:00<?, ?it/s]
10%|# | 1/10 [00:00<00:02, 3.19it/s]
20%|## | 2/10 [00:00<00:02, 3.22it/s]
30%|### | 3/10 [00:00<00:02, 3.20it/s]
40%|#### | 4/10 [00:01<00:01, 3.23it/s]
50%|##### | 5/10 [00:01<00:01, 3.24it/s]
60%|###### | 6/10 [00:01<00:01, 3.24it/s]
70%|####### | 7/10 [00:02<00:00, 3.24it/s]
80%|######## | 8/10 [00:02<00:00, 3.24it/s]
90%|######### | 9/10 [00:02<00:00, 3.23it/s]
100%|##########| 10/10 [00:03<00:00, 3.20it/s]
100%|##########| 10/10 [00:03<00:00, 3.22it/s]
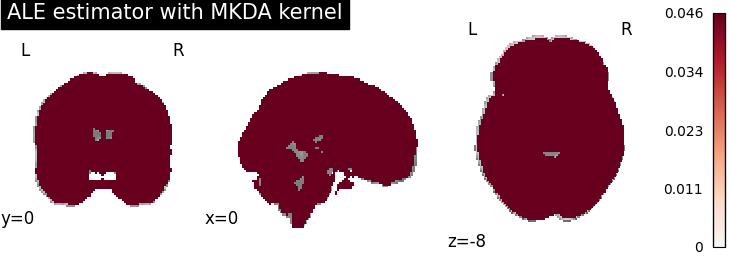
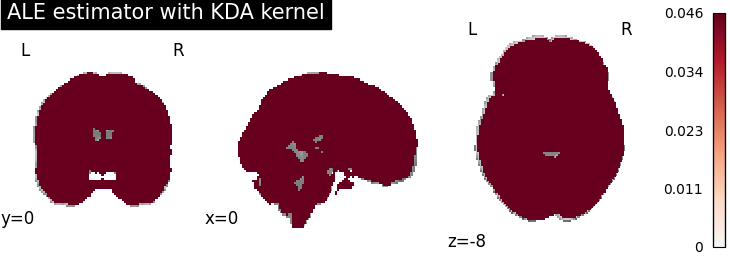
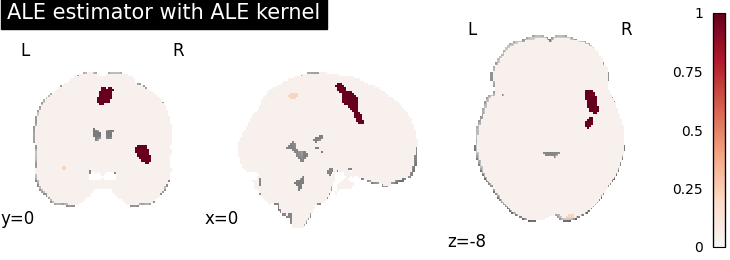
ALE
for kt_name, kt in kernel_transformers.items():
try:
ale = ALE(kernel_transformer=kt, null_method="approximate")
ale.fit(dset)
corr = FWECorrector(method="montecarlo", n_iters=10, n_cores=1)
cres = corr.transform(ale.results)
plot_stat_map(
cres.get_map("logp_desc-size_level-cluster_corr-FWE_method-montecarlo"),
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
title="ALE estimator with %s" % kt_name,
)
except AttributeError:
print(
"\nError: the %s does not currently work with the ALE meta-analysis method\n" % kt_name
)
Out:
0%| | 0/10 [00:00<?, ?it/s]
10%|# | 1/10 [00:00<00:02, 3.14it/s]
20%|## | 2/10 [00:00<00:02, 3.28it/s]
30%|### | 3/10 [00:00<00:02, 3.32it/s]
40%|#### | 4/10 [00:01<00:01, 3.33it/s]
50%|##### | 5/10 [00:01<00:01, 3.34it/s]
60%|###### | 6/10 [00:01<00:01, 3.35it/s]
70%|####### | 7/10 [00:02<00:00, 3.35it/s]
80%|######## | 8/10 [00:02<00:00, 3.37it/s]
90%|######### | 9/10 [00:02<00:00, 3.37it/s]
100%|##########| 10/10 [00:02<00:00, 3.38it/s]
100%|##########| 10/10 [00:02<00:00, 3.35it/s]
0%| | 0/10 [00:00<?, ?it/s]
10%|# | 1/10 [00:00<00:02, 3.15it/s]
20%|## | 2/10 [00:00<00:02, 3.16it/s]
30%|### | 3/10 [00:00<00:02, 3.18it/s]
40%|#### | 4/10 [00:01<00:01, 3.18it/s]
50%|##### | 5/10 [00:01<00:01, 3.18it/s]
60%|###### | 6/10 [00:01<00:01, 3.17it/s]
70%|####### | 7/10 [00:02<00:00, 3.17it/s]
80%|######## | 8/10 [00:02<00:00, 3.17it/s]
90%|######### | 9/10 [00:02<00:00, 3.16it/s]
100%|##########| 10/10 [00:03<00:00, 3.14it/s]
100%|##########| 10/10 [00:03<00:00, 3.16it/s]
0%| | 0/10 [00:00<?, ?it/s]
10%|# | 1/10 [00:00<00:02, 3.20it/s]
20%|## | 2/10 [00:00<00:02, 3.20it/s]
30%|### | 3/10 [00:00<00:02, 3.23it/s]
40%|#### | 4/10 [00:01<00:01, 3.23it/s]
50%|##### | 5/10 [00:01<00:01, 3.23it/s]
60%|###### | 6/10 [00:01<00:01, 3.20it/s]
70%|####### | 7/10 [00:02<00:00, 3.17it/s]
80%|######## | 8/10 [00:02<00:00, 3.16it/s]
90%|######### | 9/10 [00:02<00:00, 3.15it/s]
100%|##########| 10/10 [00:03<00:00, 3.15it/s]
100%|##########| 10/10 [00:03<00:00, 3.18it/s]
Total running time of the script: ( 1 minutes 45.309 seconds)