Note
Click here to download the full example code
Run image-based meta-analyses on 21 pain studies¶
Collection of NIDM-Results packs downloaded from Neurovault collection 1425, uploaded by Dr. Camille Maumet.
Caution
Dataset querying will likely change as we work to shift database querying to a remote database, rather than handling it locally with NiMARE.
import os
from nilearn.plotting import plot_stat_map
import nimare
from nimare.correct import FWECorrector
from nimare.meta import ibma
from nimare.tests.utils import get_test_data_path
Download data¶
Load Dataset¶
dset_file = os.path.join(get_test_data_path(), "nidm_pain_dset.json")
dset = nimare.dataset.Dataset(dset_file)
dset.update_path(dset_dir)
# Calculate missing images
dset.images = nimare.transforms.transform_images(
dset.images, target="z", masker=dset.masker, metadata_df=dset.metadata
)
dset.images = nimare.transforms.transform_images(
dset.images, target="varcope", masker=dset.masker, metadata_df=dset.metadata
)
Stouffer’s¶
meta = ibma.Stouffers(use_sample_size=False)
meta.fit(dset)
plot_stat_map(meta.results.get_map("z"), cut_coords=[0, 0, -8], draw_cross=False, cmap="RdBu_r")
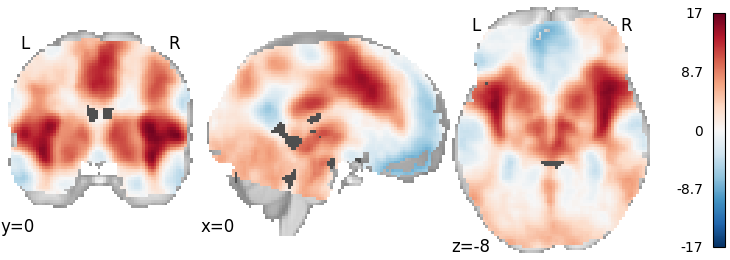
Out:
<nilearn.plotting.displays.OrthoSlicer object at 0x7fe2368ba110>
Stouffer’s with weighting by sample size¶
meta = ibma.Stouffers(use_sample_size=True)
meta.fit(dset)
plot_stat_map(meta.results.get_map("z"), cut_coords=[0, 0, -8], draw_cross=False, cmap="RdBu_r")
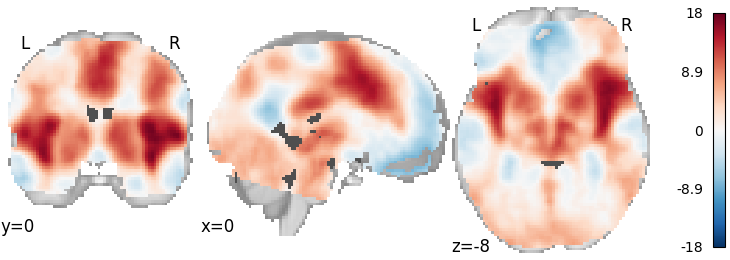
Out:
<nilearn.plotting.displays.OrthoSlicer object at 0x7fe2368d65d0>
Fisher’s¶
meta = ibma.Fishers()
meta.fit(dset)
plot_stat_map(meta.results.get_map("z"), cut_coords=[0, 0, -8], draw_cross=False, cmap="RdBu_r")
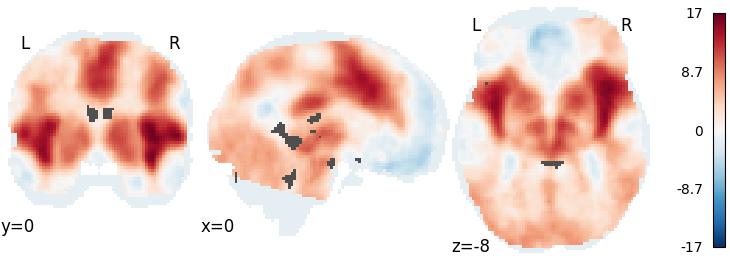
Out:
<nilearn.plotting.displays.OrthoSlicer object at 0x7fe1fd53da90>
Permuted OLS¶
meta = ibma.PermutedOLS(two_sided=True)
meta.fit(dset)
plot_stat_map(meta.results.get_map("z"), cut_coords=[0, 0, -8], draw_cross=False, cmap="RdBu_r")
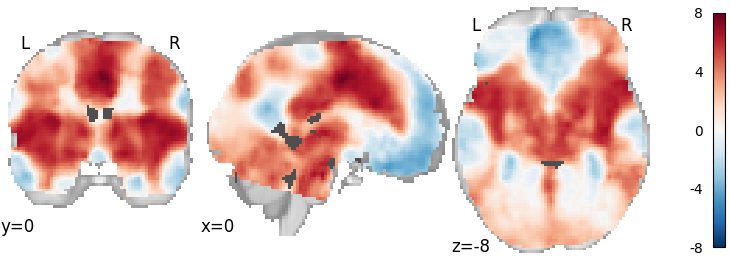
Out:
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.4/lib/python3.7/site-packages/nilearn/mass_univariate/permuted_least_squares.py:357: UserWarning: Some descriptors in 'target_vars' have zeros across all samples. These descriptors will be ignored during null distribution generation.
warnings.warn("Some descriptors in 'target_vars' have zeros across all "
<nilearn.plotting.displays.OrthoSlicer object at 0x7fe234083610>
Permuted OLS with FWE Correction¶
meta = ibma.PermutedOLS(two_sided=True)
meta.fit(dset)
corrector = FWECorrector(method="montecarlo", n_iters=100, n_cores=1)
cresult = corrector.transform(meta.results)
plot_stat_map(
cresult.get_map("z_level-voxel_corr-FWE_method-montecarlo"),
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
)
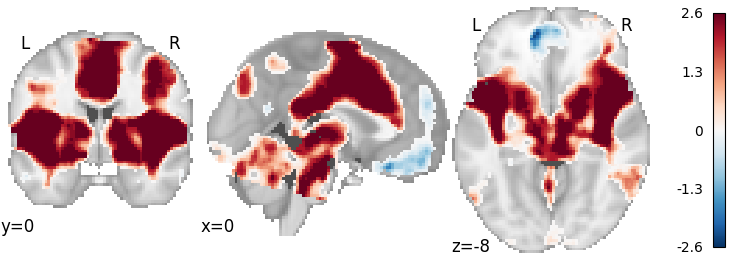
Out:
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.4/lib/python3.7/site-packages/nilearn/mass_univariate/permuted_least_squares.py:357: UserWarning: Some descriptors in 'target_vars' have zeros across all samples. These descriptors will be ignored during null distribution generation.
warnings.warn("Some descriptors in 'target_vars' have zeros across all "
<nilearn.plotting.displays.OrthoSlicer object at 0x7fe1fd8701d0>
Weighted Least Squares¶
meta = ibma.WeightedLeastSquares(tau2=0)
meta.fit(dset)
plot_stat_map(meta.results.get_map("z"), cut_coords=[0, 0, -8], draw_cross=False, cmap="RdBu_r")
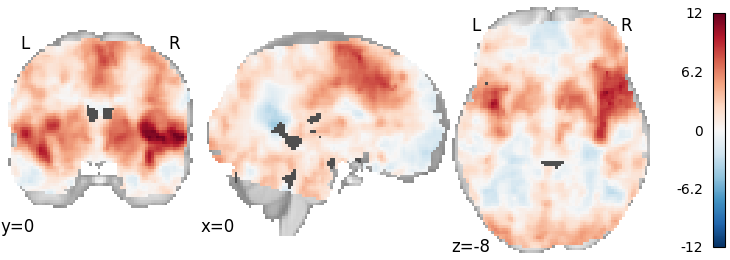
Out:
<nilearn.plotting.displays.OrthoSlicer object at 0x7fe1fd5c9c10>
DerSimonian-Laird¶
meta = ibma.DerSimonianLaird()
meta.fit(dset)
plot_stat_map(meta.results.get_map("z"), cut_coords=[0, 0, -8], draw_cross=False, cmap="RdBu_r")
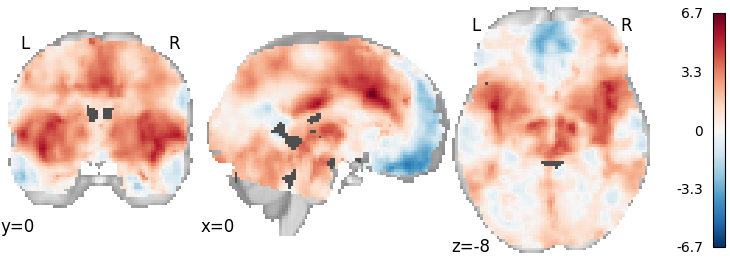
Out:
<nilearn.plotting.displays.OrthoSlicer object at 0x7fe1ea82f710>
Hedges¶
meta = ibma.Hedges()
meta.fit(dset)
plot_stat_map(meta.results.get_map("z"), cut_coords=[0, 0, -8], draw_cross=False, cmap="RdBu_r")
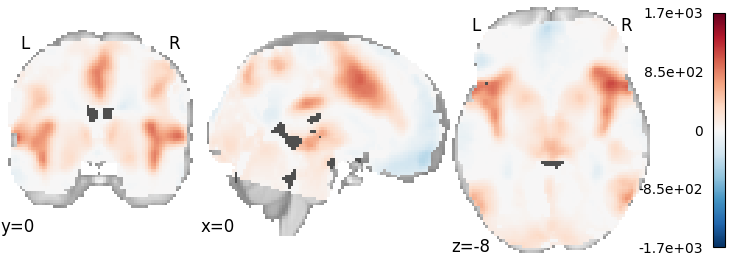
Out:
<nilearn.plotting.displays.OrthoSlicer object at 0x7fe1ea6cb350>
Total running time of the script: ( 0 minutes 50.314 seconds)