Note
Click here to download the full example code
Compare image and coordinate based meta-analyses on 21 pain studies¶
Collection of NIDM-Results packs downloaded from Neurovault collection 1425, uploaded by Dr. Camille Maumet.
import os
import pandas as pd
from nilearn.plotting import plot_stat_map
import nimare
from nimare.transforms import ImagesToCoordinates, transform_images
from nimare.meta.ibma import DerSimonianLaird
from nimare.meta.cbma import ALE
from nimare.tests.utils import get_test_data_path
Download data¶
Load Dataset¶
dset_file = os.path.join(get_test_data_path(), "nidm_pain_dset.json")
dset = nimare.dataset.Dataset(dset_file)
dset.update_path(dset_dir)
# Calculate missing statistical images from the available stats.
dset.images = transform_images(
dset.images, target="z", masker=dset.masker, metadata_df=dset.metadata
)
dset.images = nimare.transforms.transform_images(
dset.images, target="varcope", masker=dset.masker, metadata_df=dset.metadata
)
# create coordinates from statistical maps
coord_gen = ImagesToCoordinates(merge_strategy="replace")
dset = coord_gen.transform(dset)
ALE (CBMA)¶
meta_cbma = ALE()
meta_cbma.fit(dset)
plot_stat_map(
meta_cbma.results.get_map("z"), cut_coords=[0, 0, -8], draw_cross=False, cmap="RdBu_r"
)
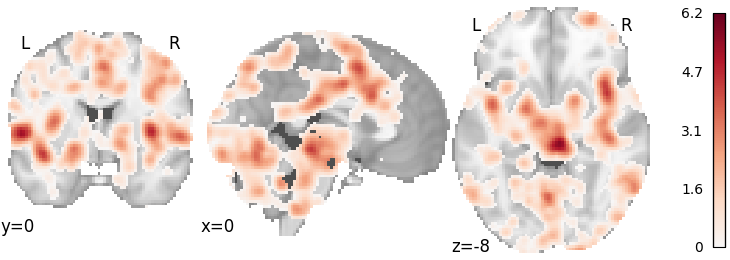
Out:
<nilearn.plotting.displays.OrthoSlicer object at 0x7f4fd4f8f150>
DerSimonian-Laird (IBMA)¶
meta_ibma = DerSimonianLaird()
meta_ibma.fit(dset)
plot_stat_map(
meta_ibma.results.get_map("z"), cut_coords=[0, 0, -8], draw_cross=False, cmap="RdBu_r"
)
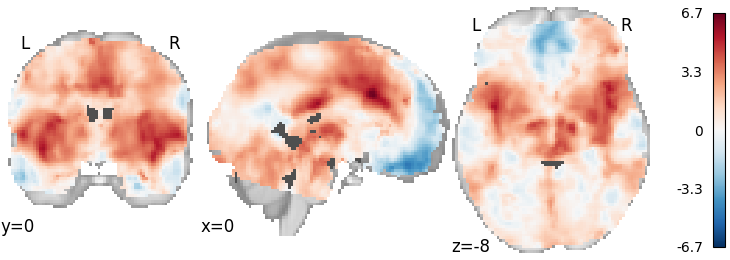
Out:
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.8/lib/python3.7/site-packages/pymare/stats.py:23: RuntimeWarning: divide by zero encountered in true_divide
w = 1. / (v + tau2)
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.8/lib/python3.7/site-packages/pymare/estimators/estimators.py:200: RuntimeWarning: divide by zero encountered in true_divide
w = 1. / v
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.8/lib/python3.7/site-packages/nilearn/_utils/niimg.py:62: UserWarning: Non-finite values detected. These values will be replaced with zeros.
"Non-finite values detected. "
<nilearn.plotting.displays.OrthoSlicer object at 0x7f4fd508b7d0>
Compare CBMA and IBMA Z-maps¶
stat_df = pd.DataFrame(
{
"CBMA": meta_cbma.results.get_map("z", return_type="array"),
"IBMA": meta_ibma.results.get_map("z", return_type="array").squeeze(),
}
)
print(stat_df.corr())
Out:
CBMA IBMA
CBMA 1.000000 0.547751
IBMA 0.547751 1.000000
Total running time of the script: ( 3 minutes 30.500 seconds)