Note
Click here to download the full example code
Image-based meta-analysis algorithms
A tour of IBMA algorithms in NiMARE.
This tutorial is intended to provide a brief description and example of each of the IBMA algorithms implemented in NiMARE. For a more detailed introduction to the elements of an image-based meta-analysis, see other stuff.
from nilearn.plotting import plot_stat_map
Download data
Note
The data used in this example come from a collection of NIDM-Results packs downloaded from Neurovault collection 1425, uploaded by Dr. Camille Maumet.
from nimare.extract import download_nidm_pain
dset_dir = download_nidm_pain()
Load Dataset
import os
from nimare.dataset import Dataset
from nimare.transforms import ImageTransformer
from nimare.utils import get_resource_path
dset_file = os.path.join(get_resource_path(), "nidm_pain_dset.json")
dset = Dataset(dset_file)
dset.update_path(dset_dir)
# Calculate missing images
xformer = ImageTransformer(target=["varcope", "z"])
dset = xformer.transform(dset)
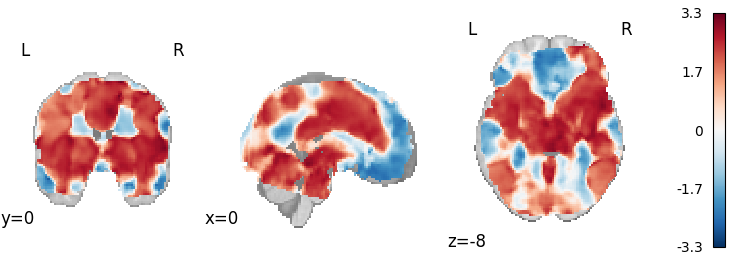
Stouffer’s
Out:
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.12/lib/python3.7/site-packages/nilearn/_utils/niimg.py:62: UserWarning: Non-finite values detected. These values will be replaced with zeros.
"Non-finite values detected. "
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7fe0a3d86b90>
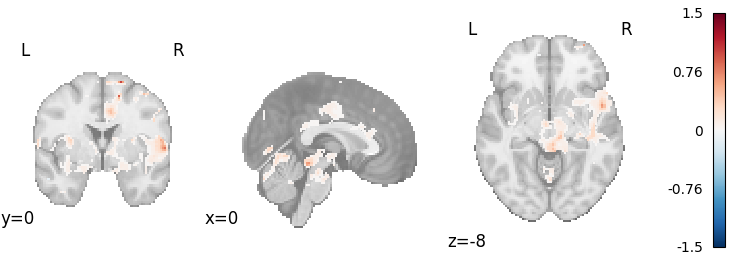
Stouffer’s with weighting by sample size
meta = Stouffers(use_sample_size=True, resample=True)
results = meta.fit(dset)
plot_stat_map(
results.get_map("z"),
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
)
Out:
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.12/lib/python3.7/site-packages/nilearn/_utils/niimg.py:62: UserWarning: Non-finite values detected. These values will be replaced with zeros.
"Non-finite values detected. "
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7fe0c33208d0>
Fisher’s
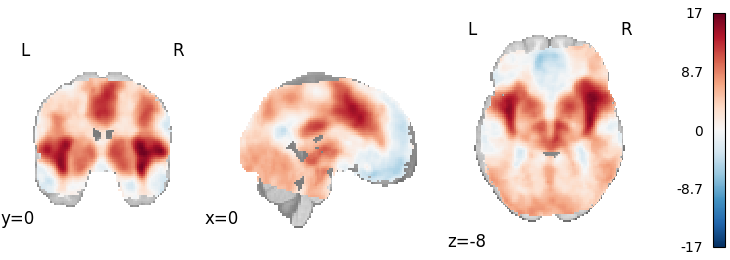
Out:
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.12/lib/python3.7/site-packages/nilearn/_utils/niimg.py:62: UserWarning: Non-finite values detected. These values will be replaced with zeros.
"Non-finite values detected. "
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7fe0c0a12910>
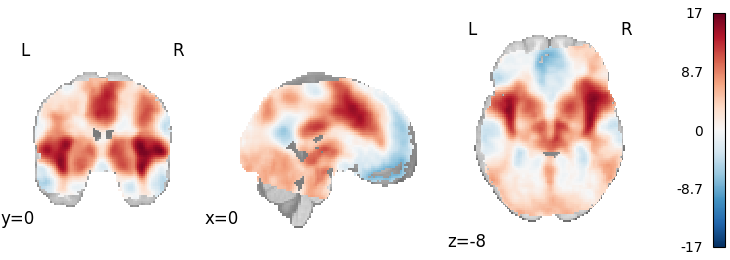
Permuted OLS
from nimare.correct import FWECorrector
from nimare.meta.ibma import PermutedOLS
meta = PermutedOLS(two_sided=True, resample=True)
results = meta.fit(dset)
plot_stat_map(
results.get_map("z"),
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
)
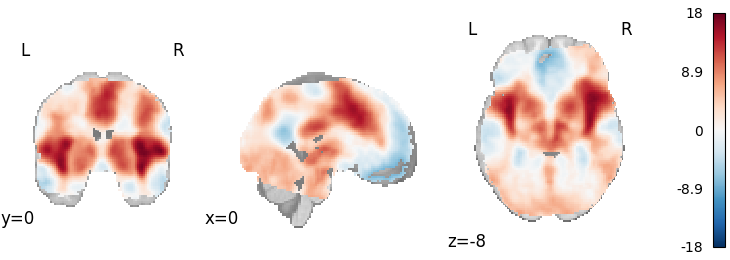
corrector = FWECorrector(method="montecarlo", n_iters=100, n_cores=1)
cresult = corrector.transform(results)
plot_stat_map(
cresult.get_map("z_level-voxel_corr-FWE_method-montecarlo"),
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
)
Out:
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.12/lib/python3.7/site-packages/nilearn/_utils/niimg.py:62: UserWarning: Non-finite values detected. These values will be replaced with zeros.
"Non-finite values detected. "
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7fe0a3ec7590>
Weighted Least Squares
from nimare.meta.ibma import WeightedLeastSquares
meta = WeightedLeastSquares(tau2=0, resample=True)
results = meta.fit(dset)
plot_stat_map(
results.get_map("z"),
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
)
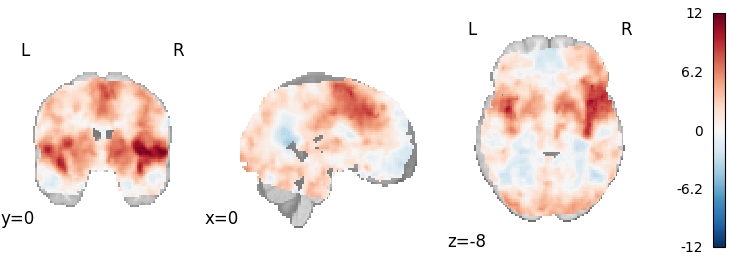
Out:
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.12/lib/python3.7/site-packages/nilearn/_utils/niimg.py:62: UserWarning: Non-finite values detected. These values will be replaced with zeros.
"Non-finite values detected. "
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7fe0a3ae0110>
DerSimonian-Laird
from nimare.meta.ibma import DerSimonianLaird
meta = DerSimonianLaird(resample=True)
results = meta.fit(dset)
plot_stat_map(
results.get_map("z"),
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
)
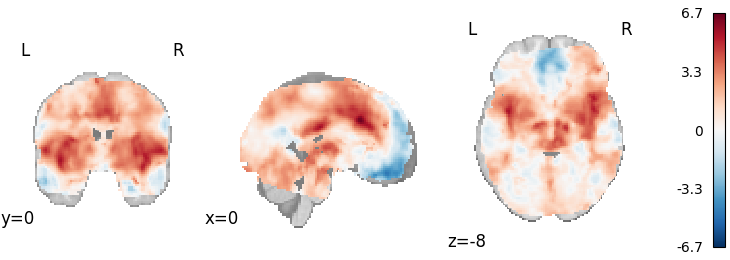
Out:
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.12/lib/python3.7/site-packages/nilearn/_utils/niimg.py:62: UserWarning: Non-finite values detected. These values will be replaced with zeros.
"Non-finite values detected. "
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7fe0a38f5b50>
Hedges
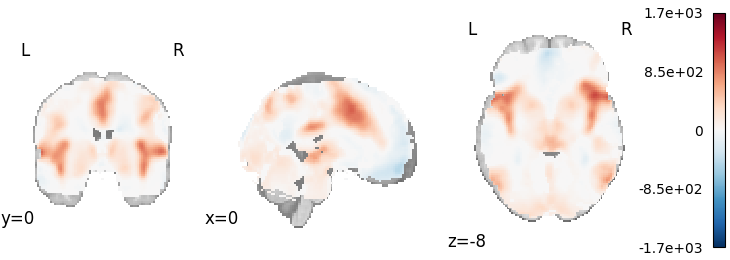
Out:
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.12/lib/python3.7/site-packages/nilearn/_utils/niimg.py:62: UserWarning: Non-finite values detected. These values will be replaced with zeros.
"Non-finite values detected. "
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7fe0c085c190>
Total running time of the script: ( 1 minutes 32.616 seconds)