Note
Go to the end to download the full example code
Discrete functional decoding
Perform meta-analytic functional decoding on regions of interest.
We can use the methods in nimare.decode.discrete to apply functional
characterization analysis to regions of interest or subsets of the Dataset.
import os
import nibabel as nib
import numpy as np
from nilearn.plotting import plot_roi
from nimare.dataset import Dataset
from nimare.decode import discrete
from nimare.utils import get_resource_path
Load dataset with abstracts
We’ll load a small dataset composed only of studies in Neurosynth with Angela Laird as a coauthor, for the sake of speed.
dset = Dataset(os.path.join(get_resource_path(), "neurosynth_laird_studies.json"))
dset.annotations.head(5)
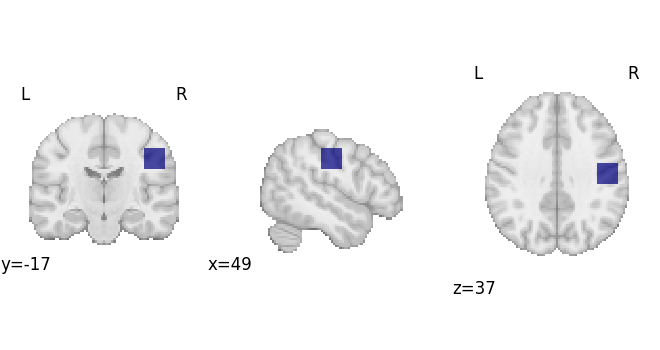
Create a region of interest
# First we'll make an ROI
arr = np.zeros(dset.masker.mask_img.shape, np.int32)
arr[65:75, 50:60, 50:60] = 1
mask_img = nib.Nifti1Image(arr, dset.masker.mask_img.affine)
plot_roi(mask_img, draw_cross=False)
# Get studies with voxels in the mask
ids = dset.get_studies_by_mask(mask_img)
Decode an ROI image using the BrainMap method
# Run the decoder
decoder = discrete.BrainMapDecoder(correction=None)
decoder.fit(dset)
decoded_df = decoder.transform(ids=ids)
decoded_df.sort_values(by="probReverse", ascending=False).head()
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/latest/lib/python3.9/site-packages/numpy/core/fromnumeric.py:86: FutureWarning:
The behavior of DataFrame.sum with axis=None is deprecated, in a future version this will reduce over both axes and return a scalar. To retain the old behavior, pass axis=0 (or do not pass axis)
Decode an ROI image using the Neurosynth chi-square method
# Run the decoder
decoder = discrete.NeurosynthDecoder(correction=None)
decoder.fit(dset)
decoded_df = decoder.transform(ids=ids)
decoded_df.sort_values(by="probReverse", ascending=False).head()
Decode an ROI image using the Neurosynth ROI association method
# This method decodes the ROI image directly, rather than comparing subsets of the Dataset like the
# other two.
decoder = discrete.ROIAssociationDecoder(mask_img)
decoder.fit(dset)
# The `transform` method doesn't take any parameters.
decoded_df = decoder.transform()
decoded_df.sort_values(by="r", ascending=False).head()
Total running time of the script: (0 minutes 3.907 seconds)