Note
Click here to download the full example code
The Corrector class
Here we take a look at multiple comparisons correction in meta-analyses.
import matplotlib.pyplot as plt
import seaborn as sns
from nilearn.plotting import plot_stat_map
Out:
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.12/lib/python3.7/site-packages/seaborn/cm.py:1582: UserWarning: Trying to register the cmap 'rocket' which already exists.
mpl_cm.register_cmap(_name, _cmap)
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.12/lib/python3.7/site-packages/seaborn/cm.py:1583: UserWarning: Trying to register the cmap 'rocket_r' which already exists.
mpl_cm.register_cmap(_name + "_r", _cmap_r)
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.12/lib/python3.7/site-packages/seaborn/cm.py:1582: UserWarning: Trying to register the cmap 'mako' which already exists.
mpl_cm.register_cmap(_name, _cmap)
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.12/lib/python3.7/site-packages/seaborn/cm.py:1583: UserWarning: Trying to register the cmap 'mako_r' which already exists.
mpl_cm.register_cmap(_name + "_r", _cmap_r)
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.12/lib/python3.7/site-packages/seaborn/cm.py:1582: UserWarning: Trying to register the cmap 'icefire' which already exists.
mpl_cm.register_cmap(_name, _cmap)
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.12/lib/python3.7/site-packages/seaborn/cm.py:1583: UserWarning: Trying to register the cmap 'icefire_r' which already exists.
mpl_cm.register_cmap(_name + "_r", _cmap_r)
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.12/lib/python3.7/site-packages/seaborn/cm.py:1582: UserWarning: Trying to register the cmap 'vlag' which already exists.
mpl_cm.register_cmap(_name, _cmap)
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.12/lib/python3.7/site-packages/seaborn/cm.py:1583: UserWarning: Trying to register the cmap 'vlag_r' which already exists.
mpl_cm.register_cmap(_name + "_r", _cmap_r)
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.12/lib/python3.7/site-packages/seaborn/cm.py:1582: UserWarning: Trying to register the cmap 'flare' which already exists.
mpl_cm.register_cmap(_name, _cmap)
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.12/lib/python3.7/site-packages/seaborn/cm.py:1583: UserWarning: Trying to register the cmap 'flare_r' which already exists.
mpl_cm.register_cmap(_name + "_r", _cmap_r)
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.12/lib/python3.7/site-packages/seaborn/cm.py:1582: UserWarning: Trying to register the cmap 'crest' which already exists.
mpl_cm.register_cmap(_name, _cmap)
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.12/lib/python3.7/site-packages/seaborn/cm.py:1583: UserWarning: Trying to register the cmap 'crest_r' which already exists.
mpl_cm.register_cmap(_name + "_r", _cmap_r)
Download data
from nimare.extract import download_nidm_pain
dset_dir = download_nidm_pain()
Load Dataset
import os
from nimare.dataset import Dataset
from nimare.utils import get_resource_path
dset_file = os.path.join(get_resource_path(), "nidm_pain_dset.json")
dset = Dataset(dset_file)
dset.update_path(dset_dir)
mask_img = dset.masker.mask_img
Multiple comparisons correction in coordinate-based meta-analyses
Tip
For more information multiple comparisons correction and CBMA in NiMARE, see Multiple comparisons correction.
from nimare.meta.cbma.ale import ALE
# First, we need to fit the Estimator to the Dataset.
meta = ALE(null_method="approximate")
results = meta.fit(dset)
# We can check which FWE correction methods are available for the ALE Estimator
# with the ``inspect`` class method.
from nimare.correct import FWECorrector
print(FWECorrector.inspect(results))
Out:
['bonferroni', 'montecarlo']
Apply the Corrector to the MetaResult
Now that we know what FWE correction methods are available, we can use one.
The “montecarlo” method is a special one that is implemented within the Estimator, rather than in the Corrector.
corr = FWECorrector(method="montecarlo", n_iters=50, n_cores=2)
cres = corr.transform(results)
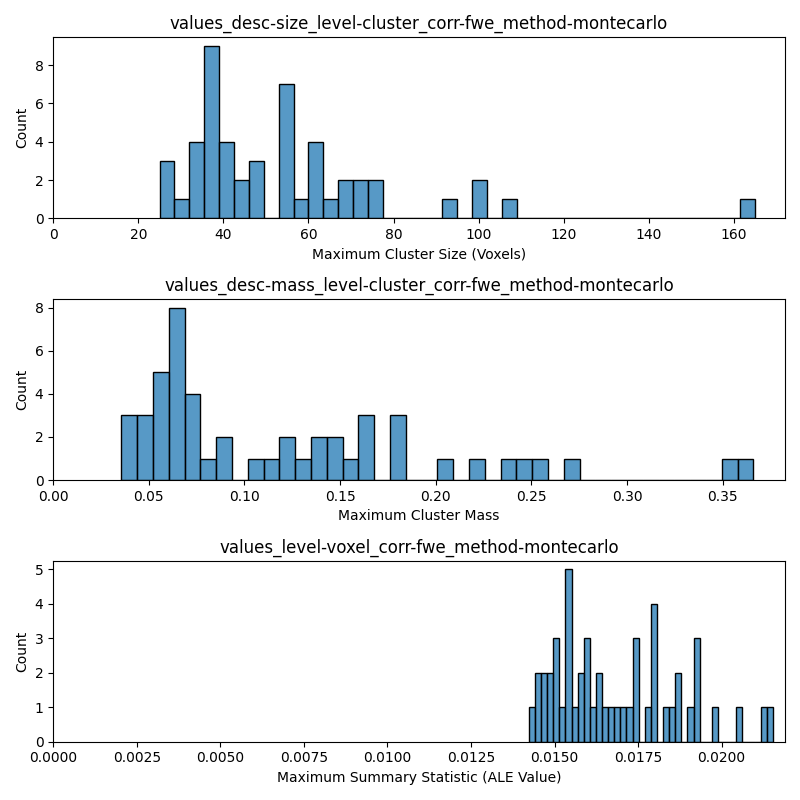
DISTS_TO_PLOT = [
"values_desc-size_level-cluster_corr-fwe_method-montecarlo",
"values_desc-mass_level-cluster_corr-fwe_method-montecarlo",
"values_level-voxel_corr-fwe_method-montecarlo",
]
XLABELS = [
"Maximum Cluster Size (Voxels)",
"Maximum Cluster Mass",
"Maximum Summary Statistic (ALE Value)",
]
fig, axes = plt.subplots(figsize=(8, 8), nrows=3)
null_dists = cres.estimator.null_distributions_
for i_ax, dist_name in enumerate(DISTS_TO_PLOT):
xlabel = XLABELS[i_ax]
sns.histplot(x=null_dists[dist_name], bins=40, ax=axes[i_ax])
axes[i_ax].set_title(dist_name)
axes[i_ax].set_xlabel(xlabel)
axes[i_ax].set_xlim(0, None)
fig.tight_layout()
Out:
0%| | 0/50 [00:00<?, ?it/s]
2%|2 | 1/50 [00:05<04:18, 5.27s/it]
4%|4 | 2/50 [00:05<01:47, 2.24s/it]
6%|6 | 3/50 [00:06<01:16, 1.62s/it]
8%|8 | 4/50 [00:06<00:47, 1.04s/it]
10%|# | 5/50 [00:07<00:41, 1.07it/s]
12%|#2 | 6/50 [00:07<00:29, 1.49it/s]
14%|#4 | 7/50 [00:08<00:31, 1.37it/s]
16%|#6 | 8/50 [00:08<00:23, 1.81it/s]
18%|#8 | 9/50 [00:09<00:24, 1.67it/s]
20%|## | 10/50 [00:09<00:18, 2.13it/s]
22%|##2 | 11/50 [00:10<00:22, 1.73it/s]
24%|##4 | 12/50 [00:10<00:17, 2.17it/s]
26%|##6 | 13/50 [00:10<00:19, 1.87it/s]
28%|##8 | 14/50 [00:11<00:15, 2.33it/s]
30%|### | 15/50 [00:11<00:19, 1.82it/s]
32%|###2 | 16/50 [00:12<00:15, 2.20it/s]
34%|###4 | 17/50 [00:12<00:16, 1.97it/s]
36%|###6 | 18/50 [00:13<00:13, 2.30it/s]
38%|###8 | 19/50 [00:13<00:16, 1.91it/s]
40%|#### | 20/50 [00:14<00:13, 2.22it/s]
42%|####2 | 21/50 [00:14<00:14, 2.01it/s]
44%|####4 | 22/50 [00:14<00:12, 2.32it/s]
46%|####6 | 23/50 [00:15<00:14, 1.90it/s]
48%|####8 | 24/50 [00:15<00:11, 2.24it/s]
50%|##### | 25/50 [00:16<00:12, 1.97it/s]
52%|#####2 | 26/50 [00:16<00:10, 2.34it/s]
54%|#####4 | 27/50 [00:17<00:12, 1.87it/s]
56%|#####6 | 28/50 [00:17<00:10, 2.17it/s]
58%|#####8 | 29/50 [00:18<00:10, 1.99it/s]
60%|###### | 30/50 [00:18<00:08, 2.28it/s]
62%|######2 | 31/50 [00:19<00:09, 1.93it/s]
64%|######4 | 32/50 [00:19<00:08, 2.15it/s]
66%|######6 | 33/50 [00:20<00:08, 2.02it/s]
68%|######8 | 34/50 [00:20<00:07, 2.27it/s]
70%|####### | 35/50 [00:21<00:07, 1.97it/s]
72%|#######2 | 36/50 [00:21<00:06, 2.20it/s]
74%|#######4 | 37/50 [00:22<00:06, 2.12it/s]
76%|#######6 | 38/50 [00:22<00:05, 2.23it/s]
78%|#######8 | 39/50 [00:23<00:05, 2.06it/s]
80%|######## | 40/50 [00:23<00:04, 2.11it/s]
82%|########2 | 41/50 [00:24<00:04, 2.18it/s]
84%|########4 | 42/50 [00:24<00:03, 2.19it/s]
86%|########6 | 43/50 [00:25<00:03, 2.03it/s]
88%|########8 | 44/50 [00:25<00:02, 2.09it/s]
90%|######### | 45/50 [00:26<00:02, 2.14it/s]
92%|#########2| 46/50 [00:26<00:01, 2.18it/s]
94%|#########3| 47/50 [00:27<00:01, 2.03it/s]
96%|#########6| 48/50 [00:27<00:00, 2.07it/s]
98%|#########8| 49/50 [00:27<00:00, 2.19it/s]
100%|##########| 50/50 [00:28<00:00, 2.36it/s]
100%|##########| 50/50 [00:28<00:00, 1.76it/s]
Show corrected results
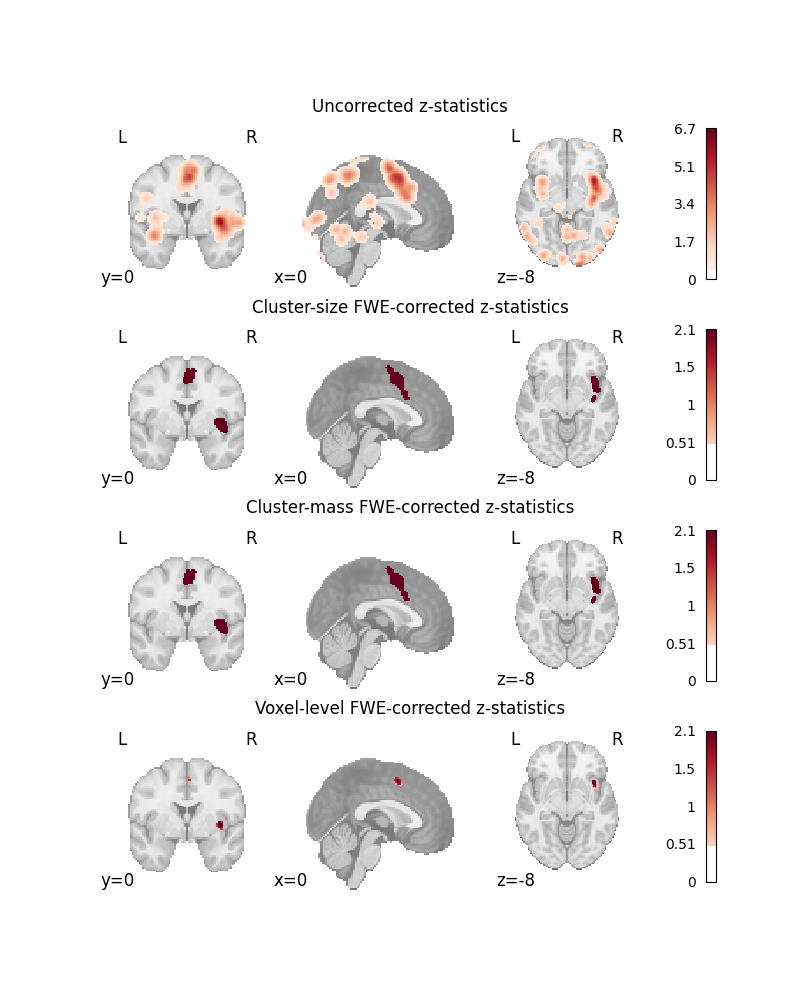
MAPS_TO_PLOT = [
"z",
"z_desc-size_level-cluster_corr-FWE_method-montecarlo",
"z_desc-mass_level-cluster_corr-FWE_method-montecarlo",
"z_level-voxel_corr-FWE_method-montecarlo",
]
TITLES = [
"Uncorrected z-statistics",
"Cluster-size FWE-corrected z-statistics",
"Cluster-mass FWE-corrected z-statistics",
"Voxel-level FWE-corrected z-statistics",
]
fig, axes = plt.subplots(figsize=(8, 10), nrows=4)
for i_ax, map_name in enumerate(MAPS_TO_PLOT):
title = TITLES[i_ax]
plot_stat_map(
cres.get_map(map_name),
draw_cross=False,
cmap="RdBu_r",
threshold=0.5,
cut_coords=[0, 0, -8],
figure=fig,
axes=axes[i_ax],
)
axes[i_ax].set_title(title)
Multiple comparisons correction in image-based meta-analyses
from nimare.correct import FDRCorrector
from nimare.meta.ibma import Stouffers
meta = Stouffers(resample=True)
results = meta.fit(dset)
print(f"FWECorrector options: {FWECorrector.inspect(results)}")
print(f"FDRCorrector options: {FDRCorrector.inspect(results)}")
Out:
FWECorrector options: ['bonferroni']
FDRCorrector options: ['indep', 'negcorr']
Note that the FWECorrector does not support a “montecarlo” method for the Stouffers Estimator. This is because NiMARE does not have a Monte Carlo-based method implemented for most IBMA algorithms.
Apply the Corrector to the MetaResult
corr = FDRCorrector(method="indep", alpha=0.05)
cres = corr.transform(results)
Show corrected results
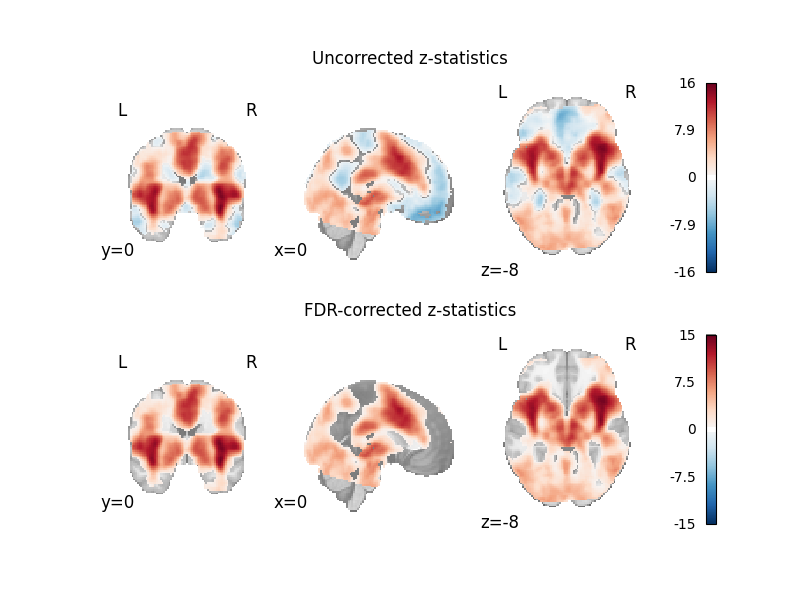
fig, axes = plt.subplots(figsize=(8, 6), nrows=2)
plot_stat_map(
cres.get_map("z"),
draw_cross=False,
cmap="RdBu_r",
threshold=0.5,
cut_coords=[0, 0, -8],
figure=fig,
axes=axes[0],
)
axes[0].set_title("Uncorrected z-statistics")
plot_stat_map(
cres.get_map("z_corr-FDR_method-indep"),
draw_cross=False,
cmap="RdBu_r",
threshold=0.5,
cut_coords=[0, 0, -8],
figure=fig,
axes=axes[1],
)
axes[1].set_title("FDR-corrected z-statistics")
Out:
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/0.0.12/lib/python3.7/site-packages/nilearn/_utils/niimg.py:62: UserWarning: Non-finite values detected. These values will be replaced with zeros.
"Non-finite values detected. "
Text(0.5, 1.0, 'FDR-corrected z-statistics')
Total running time of the script: ( 0 minutes 44.064 seconds)