nimare.meta.cbma.base.CBMAEstimator
- class CBMAEstimator(kernel_transformer, *, mask=None, **kwargs)[source]
Bases:
EstimatorBase class for coordinate-based meta-analysis methods.
Changed in version 0.0.12:
Remove low_memory option
CBMA-specific elements of
MetaEstimatorexcised and moved intoCBMAEstimator.Generic kwargs and args converted to named kwargs. All remaining kwargs are for kernels.
Use a 4D sparse array for modeled activation maps.
Changed in version 0.0.8:
[REF] Use saved MA maps, when available.
[REF] Add low_memory option.
New in version 0.0.3.
- Parameters
kernel_transformer (
KernelTransformer, optional) – Kernel with which to convolve coordinates from dataset. Default is ALEKernel.*args – Optional arguments to the
MetaEstimator__init__ (called automatically).**kwargs – Optional keyword arguments to the
MetaEstimator__init__ (called automatically).
Methods
correct_fwe_montecarlo(result[, ...])Perform FWE correction using the max-value permutation method.
fit(dataset[, drop_invalid])Fit Estimator to Dataset.
get_params([deep])Get parameters for this estimator.
load(filename[, compressed])Load a pickled class instance from file.
save(filename[, compress])Pickle the class instance to the provided file.
set_params(**params)Set the parameters of this estimator.
- correct_fwe_montecarlo(result, voxel_thresh=0.001, n_iters=10000, n_cores=1, vfwe_only=False)[source]
Perform FWE correction using the max-value permutation method.
Only call this method from within a Corrector.
Changed in version 0.0.12:
Fix the
vfwe_onlyoption.
Changed in version 0.0.11:
Rename
*_level-clustermaps to*_desc-size_level-cluster.Add new
*_desc-mass_level-clustermaps that use cluster mass-based inference.
- Parameters
result (
MetaResult) – Result object from a CBMA meta-analysis.voxel_thresh (
float, optional) – Cluster-defining p-value threshold. Default is 0.001.n_iters (
int, optional) – Number of iterations to build the voxel-level, cluster-size, and cluster-mass FWE null distributions. Default is 10000.n_cores (
int, optional) – Number of cores to use for parallelization. If <=0, defaults to using all available cores. Default is 1.vfwe_only (
bool, optional) – If True, only calculate the voxel-level FWE-corrected maps. Voxel-level correction can be performed very quickly if the Estimator’snull_methodwas “montecarlo”. Default is False.
- Returns
images – Dictionary of 1D arrays corresponding to masked images generated by the correction procedure. The following arrays are generated by this method:
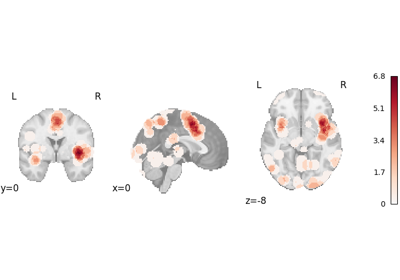
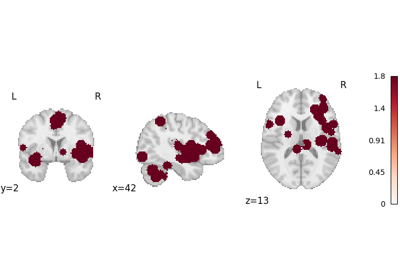
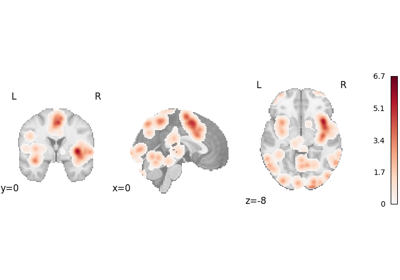
logp_desc-size_level-cluster: Cluster-level FWE-corrected-log10(p)map based on cluster size. This was previously simply called “logp_level-cluster”. This array is not generated ifvfwe_onlyisTrue.logp_desc-mass_level-cluster: Cluster-level FWE-corrected-log10(p)map based on cluster mass. According to Bullmore et al.1 and Zhang et al.2, cluster mass-based inference is more powerful than cluster size. This array is not generated ifvfwe_onlyisTrue.logp_level-voxel: Voxel-level FWE-corrected-log10(p)map. Voxel-level correction is generally more conservative than cluster-level correction, so it is only recommended for very large meta-analyses (i.e., hundreds of studies), per Eickhoff et al.3.
- Return type
Notes
If
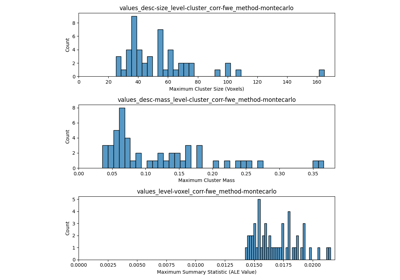
vfwe_onlyisFalse, this method adds three new keys to thenull_distributions_attribute:values_level-voxel_corr-fwe_method-montecarlo: The maximum summary statistic value from each Monte Carlo iteration. An array of shape (n_iters,).values_desc-size_level-cluster_corr-fwe_method-montecarlo: The maximum cluster size from each Monte Carlo iteration. An array of shape (n_iters,).values_desc-mass_level-cluster_corr-fwe_method-montecarlo: The maximum cluster mass from each Monte Carlo iteration. An array of shape (n_iters,).
See also
nimare.correct.FWECorrectorThe Corrector from which to call this method.
References
- 1
Edward T Bullmore, John Suckling, Stephan Overmeyer, Sophia Rabe-Hesketh, Eric Taylor, and Michael J Brammer. Global, voxel, and cluster tests, by theory and permutation, for a difference between two groups of structural mr images of the brain. IEEE transactions on medical imaging, 18(1):32–42, 1999. URL: https://doi.org/10.1109/42.750253, doi:10.1109/42.750253.
- 2
Hui Zhang, Thomas E Nichols, and Timothy D Johnson. Cluster mass inference via random field theory. Neuroimage, 44(1):51–61, 2009. URL: https://doi.org/10.1016/j.neuroimage.2008.08.017, doi:10.1016/j.neuroimage.2008.08.017.
- 3
Simon B Eickhoff, Thomas E Nichols, Angela R Laird, Felix Hoffstaedter, Katrin Amunts, Peter T Fox, Danilo Bzdok, and Claudia R Eickhoff. Behavior, sensitivity, and power of activation likelihood estimation characterized by massive empirical simulation. Neuroimage, 137:70–85, 2016. URL: https://doi.org/10.1016/j.neuroimage.2016.04.072, doi:10.1016/j.neuroimage.2016.04.072.
Examples
>>> meta = MKDADensity() >>> result = meta.fit(dset) >>> corrector = FWECorrector(method='montecarlo', voxel_thresh=0.01, n_iters=5, n_cores=1) >>> cresult = corrector.transform(result)
- fit(dataset, drop_invalid=True)[source]
Fit Estimator to Dataset.
- Parameters
- Returns
Results of Estimator fitting.
- Return type
- Variables
inputs (
dict) – Inputs used in _fit.
- classmethod load(filename, compressed=True)[source]
Load a pickled class instance from file.
- Parameters
- Returns
obj – Loaded class object.
- Return type
class object
- set_params(**params)[source]
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as pipelines). The latter have parameters of the form
<component>__<parameter>so that it’s possible to update each component of a nested object.- Return type
self