Note
Go to the end to download the full example code
Coordinate-based meta-analysis algorithms
A tour of CBMA algorithms in NiMARE.
This tutorial is intended to provide a brief description and example of each of the CBMA algorithms implemented in NiMARE. For a more detailed introduction to the elements of a coordinate-based meta-analysis, see other stuff.
Load Dataset
Note
The data used in this example come from a collection of NIDM-Results packs downloaded from Neurovault collection 1425, uploaded by Dr. Camille Maumet.
Creation of the Dataset from the NIDM-Results packs was done with custom code. The Results packs for collection 1425 are not completely NIDM-Results-compliant, so the nidmresults library could not be used to facilitate data extraction.
import os
from pprint import pprint
from nilearn.plotting import plot_stat_map
from nimare.correct import FWECorrector
from nimare.dataset import Dataset
from nimare.utils import get_resource_path
dset_file = os.path.join(get_resource_path(), "nidm_pain_dset.json")
dset = Dataset(dset_file)
# Some of the CBMA algorithms compare two Datasets,
# so we'll split this example Dataset in half.
dset1 = dset.slice(dset.ids[:10])
dset2 = dset.slice(dset.ids[10:])
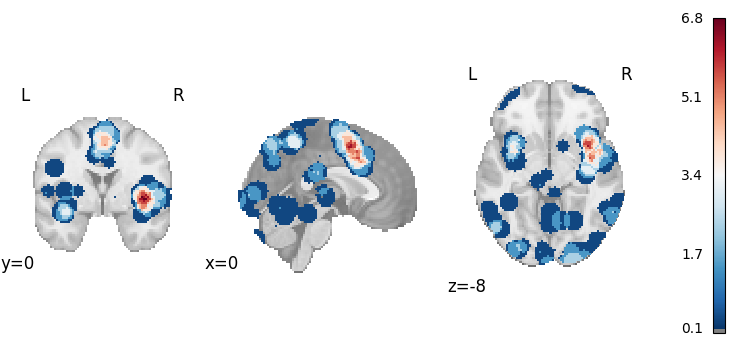
Multilevel Kernel Density Analysis
from nimare.meta.cbma.mkda import MKDADensity
meta = MKDADensity()
results = meta.fit(dset)
corr = FWECorrector(method="montecarlo", n_iters=10, n_cores=1)
cres = corr.transform(results)
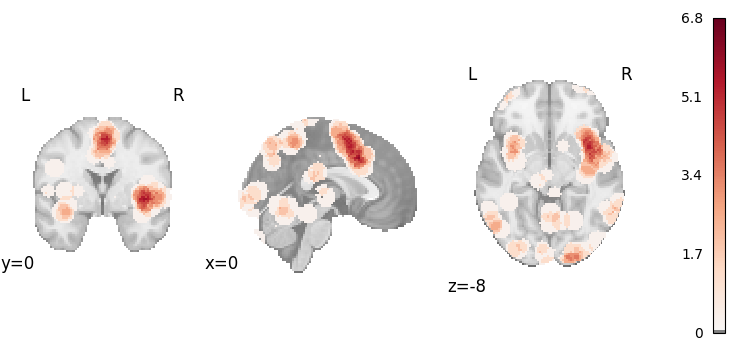
plot_stat_map(
results.get_map("z"),
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
threshold=0.1,
)
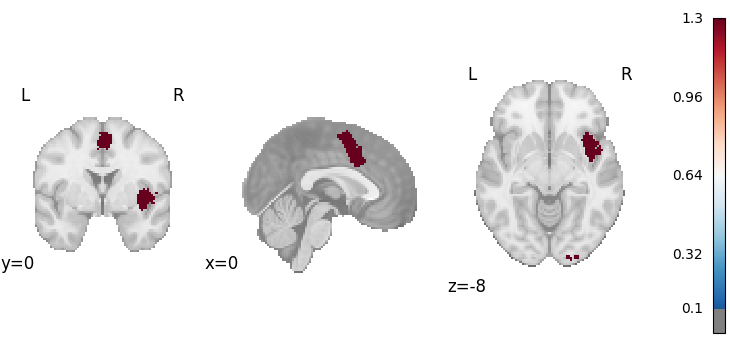
plot_stat_map(
cres.get_map("z_level-voxel_corr-FWE_method-montecarlo"),
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
threshold=0.1,
)
print("Description:")
pprint(results.description_)
print("References:")
pprint(results.bibtex_)
0%| | 0/10 [00:00<?, ?it/s]
10%|█ | 1/10 [00:00<00:02, 3.13it/s]
20%|██ | 2/10 [00:00<00:02, 3.20it/s]
30%|███ | 3/10 [00:00<00:02, 3.22it/s]
40%|████ | 4/10 [00:01<00:01, 3.22it/s]
50%|█████ | 5/10 [00:01<00:01, 3.22it/s]
60%|██████ | 6/10 [00:01<00:01, 3.23it/s]
70%|███████ | 7/10 [00:02<00:00, 3.23it/s]
80%|████████ | 8/10 [00:02<00:00, 3.22it/s]
90%|█████████ | 9/10 [00:02<00:00, 3.22it/s]
100%|██████████| 10/10 [00:03<00:00, 3.22it/s]
100%|██████████| 10/10 [00:03<00:00, 3.22it/s]
Description:
('A multilevel kernel density (MKDA) meta-analysis \\citep{wager2007meta} was '
'performed was performed with NiMARE 0.2.2+2.gbf7df33.dirty (RRID:SCR_017398; '
'\\citealt{Salo2023}), using a(n) MKDA kernel. An MKDA kernel '
'\\citep{wager2007meta} was used to generate study-wise modeled activation '
'maps from coordinates. In this kernel method, each coordinate is convolved '
'with a sphere with a radius of 10.0 and a value of 1. For voxels with '
'overlapping spheres, the maximum value was retained. Summary statistics (OF '
'values) were converted to p-values using an approximate null distribution. '
'The input dataset included 267 foci from 21 experiments.')
References:
('@article{Salo2023,\n'
' doi = {10.52294/001c.87681},\n'
' url = {https://doi.org/10.52294/001c.87681},\n'
' year = {2023},\n'
' volume = {3},\n'
' pages = {1 - 32},\n'
' author = {Taylor Salo and Tal Yarkoni and Thomas E. Nichols and '
'Jean-Baptiste Poline and Murat Bilgel and Katherine L. Bottenhorn and Dorota '
'Jarecka and James D. Kent and Adam Kimbler and Dylan M. Nielson and Kendra '
'M. Oudyk and Julio A. Peraza and Alexandre Pérez and Puck C. Reeders and '
'Julio A. Yanes and Angela R. Laird},\n'
' title = {NiMARE: Neuroimaging Meta-Analysis Research Environment},\n'
' journal = {Aperture Neuro}\n'
'}\n'
'@article{wager2007meta,\n'
' title={Meta-analysis of functional neuroimaging data: current and future '
'directions},\n'
' author={Wager, Tor D and Lindquist, Martin and Kaplan, Lauren},\n'
' journal={Social cognitive and affective neuroscience},\n'
' volume={2},\n'
' number={2},\n'
' pages={150--158},\n'
' year={2007},\n'
' publisher={Oxford University Press},\n'
' url={https://doi.org/10.1093/scan/nsm015},\n'
' doi={10.1093/scan/nsm015}\n'
'}')
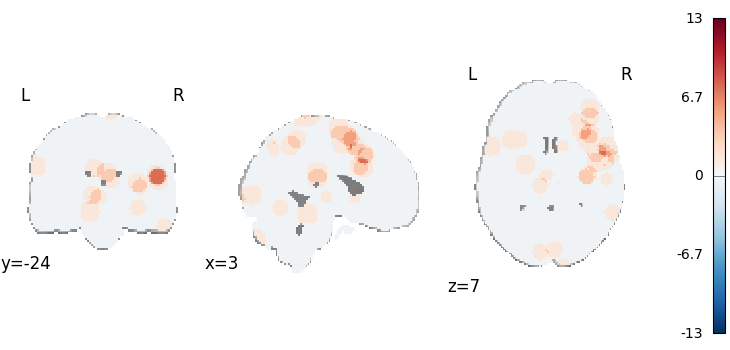
MKDA Chi-Squared
from nimare.meta.cbma.mkda import MKDAChi2
meta = MKDAChi2(kernel__r=10)
results = meta.fit(dset1, dset2)
corr = FWECorrector(method="montecarlo", n_iters=10, n_cores=1)
cres = corr.transform(results)
plot_stat_map(
results.get_map("z_desc-uniformity"),
draw_cross=False,
cmap="RdBu_r",
threshold=0.1,
)
plot_stat_map(
cres.get_map("z_desc-uniformitySize_level-cluster_corr-FWE_method-montecarlo"),
draw_cross=False,
cmap="RdBu_r",
threshold=0.1,
)
print("Description:")
pprint(results.description_)
print("References:")
pprint(results.bibtex_)
/home/docs/checkouts/readthedocs.org/user_builds/nimare/checkouts/latest/nimare/meta/cbma/mkda.py:473: RuntimeWarning: invalid value encountered in divide
pFgA = pAgF * pF / pA
/home/docs/checkouts/readthedocs.org/user_builds/nimare/checkouts/latest/nimare/meta/cbma/mkda.py:480: RuntimeWarning: invalid value encountered in divide
pFgA_prior = pAgF * self.prior / pAgF_prior
0%| | 0/10 [00:00<?, ?it/s]
10%|█ | 1/10 [00:00<00:05, 1.56it/s]
20%|██ | 2/10 [00:01<00:05, 1.58it/s]
30%|███ | 3/10 [00:01<00:04, 1.58it/s]
40%|████ | 4/10 [00:02<00:03, 1.57it/s]
50%|█████ | 5/10 [00:03<00:03, 1.57it/s]
60%|██████ | 6/10 [00:03<00:02, 1.57it/s]
70%|███████ | 7/10 [00:04<00:01, 1.57it/s]
80%|████████ | 8/10 [00:05<00:01, 1.55it/s]
90%|█████████ | 9/10 [00:05<00:00, 1.55it/s]
100%|██████████| 10/10 [00:06<00:00, 1.55it/s]
100%|██████████| 10/10 [00:06<00:00, 1.56it/s]
Description:
('A multilevel kernel density chi-squared analysis \\citep{wager2007meta} was '
'performed according to the same procedure as implemented in Neurosynth with '
'NiMARE 0.2.2+2.gbf7df33.dirty (RRID:SCR_017398; \\citealt{Salo2023}), using '
'a(n) MKDA kernel. An MKDA kernel \\citep{wager2007meta} was used to generate '
'study-wise modeled activation maps from coordinates. In this kernel method, '
'each coordinate is convolved with a sphere with a radius of 10.0 and a value '
'of 1. For voxels with overlapping spheres, the maximum value was retained. '
'This analysis calculated several measures. The first dataset was evaluated '
'for uniformity of activation via a one-way chi-square test. The first input '
'dataset included 147 foci from 10 experiments. The second input dataset '
'included 120 foci from 11 experiments.')
References:
('@article{Salo2023,\n'
' doi = {10.52294/001c.87681},\n'
' url = {https://doi.org/10.52294/001c.87681},\n'
' year = {2023},\n'
' volume = {3},\n'
' pages = {1 - 32},\n'
' author = {Taylor Salo and Tal Yarkoni and Thomas E. Nichols and '
'Jean-Baptiste Poline and Murat Bilgel and Katherine L. Bottenhorn and Dorota '
'Jarecka and James D. Kent and Adam Kimbler and Dylan M. Nielson and Kendra '
'M. Oudyk and Julio A. Peraza and Alexandre Pérez and Puck C. Reeders and '
'Julio A. Yanes and Angela R. Laird},\n'
' title = {NiMARE: Neuroimaging Meta-Analysis Research Environment},\n'
' journal = {Aperture Neuro}\n'
'}\n'
'@article{wager2007meta,\n'
' title={Meta-analysis of functional neuroimaging data: current and future '
'directions},\n'
' author={Wager, Tor D and Lindquist, Martin and Kaplan, Lauren},\n'
' journal={Social cognitive and affective neuroscience},\n'
' volume={2},\n'
' number={2},\n'
' pages={150--158},\n'
' year={2007},\n'
' publisher={Oxford University Press},\n'
' url={https://doi.org/10.1093/scan/nsm015},\n'
' doi={10.1093/scan/nsm015}\n'
'}')
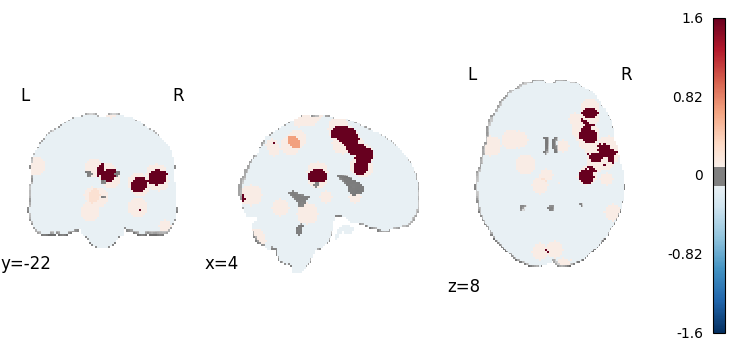
Kernel Density Analysis
from nimare.meta.cbma.mkda import KDA
meta = KDA()
results = meta.fit(dset)
corr = FWECorrector(method="montecarlo", n_iters=10, n_cores=1)
cres = corr.transform(results)
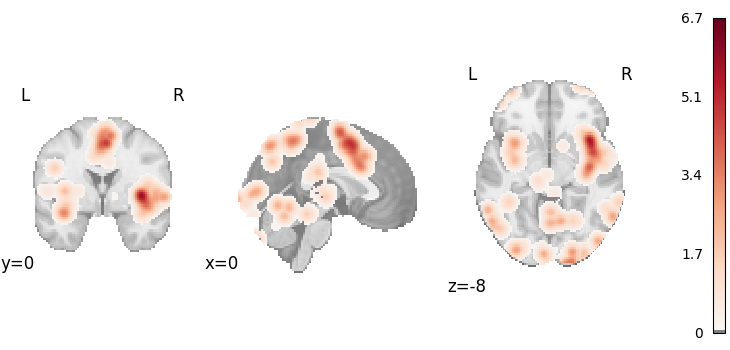
plot_stat_map(
results.get_map("z"),
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
threshold=0.1,
)
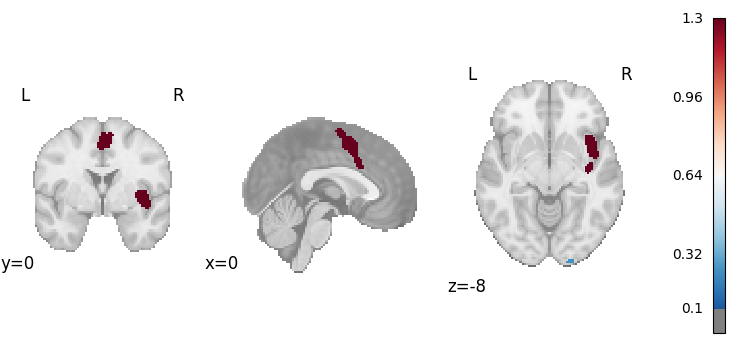
plot_stat_map(
cres.get_map("z_desc-size_level-cluster_corr-FWE_method-montecarlo"),
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
threshold=0.1,
)
print("Description:")
pprint(results.description_)
print("References:")
pprint(results.bibtex_)
0%| | 0/10 [00:00<?, ?it/s]/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/latest/lib/python3.9/site-packages/nilearn/masking.py:974: UserWarning: Data array used to create a new image contains 64-bit ints. This is likely due to creating the array with numpy and passing `int` as the `dtype`. Many tools such as FSL and SPM cannot deal with int64 in Nifti images, so for compatibility the data has been converted to int32.
return new_img_like(mask_img, unmasked, affine)
10%|█ | 1/10 [00:00<00:03, 2.95it/s]/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/latest/lib/python3.9/site-packages/nilearn/masking.py:974: UserWarning: Data array used to create a new image contains 64-bit ints. This is likely due to creating the array with numpy and passing `int` as the `dtype`. Many tools such as FSL and SPM cannot deal with int64 in Nifti images, so for compatibility the data has been converted to int32.
return new_img_like(mask_img, unmasked, affine)
20%|██ | 2/10 [00:00<00:02, 3.03it/s]/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/latest/lib/python3.9/site-packages/nilearn/masking.py:974: UserWarning: Data array used to create a new image contains 64-bit ints. This is likely due to creating the array with numpy and passing `int` as the `dtype`. Many tools such as FSL and SPM cannot deal with int64 in Nifti images, so for compatibility the data has been converted to int32.
return new_img_like(mask_img, unmasked, affine)
30%|███ | 3/10 [00:00<00:02, 3.07it/s]/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/latest/lib/python3.9/site-packages/nilearn/masking.py:974: UserWarning: Data array used to create a new image contains 64-bit ints. This is likely due to creating the array with numpy and passing `int` as the `dtype`. Many tools such as FSL and SPM cannot deal with int64 in Nifti images, so for compatibility the data has been converted to int32.
return new_img_like(mask_img, unmasked, affine)
40%|████ | 4/10 [00:01<00:01, 3.10it/s]/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/latest/lib/python3.9/site-packages/nilearn/masking.py:974: UserWarning: Data array used to create a new image contains 64-bit ints. This is likely due to creating the array with numpy and passing `int` as the `dtype`. Many tools such as FSL and SPM cannot deal with int64 in Nifti images, so for compatibility the data has been converted to int32.
return new_img_like(mask_img, unmasked, affine)
50%|█████ | 5/10 [00:01<00:01, 3.12it/s]/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/latest/lib/python3.9/site-packages/nilearn/masking.py:974: UserWarning: Data array used to create a new image contains 64-bit ints. This is likely due to creating the array with numpy and passing `int` as the `dtype`. Many tools such as FSL and SPM cannot deal with int64 in Nifti images, so for compatibility the data has been converted to int32.
return new_img_like(mask_img, unmasked, affine)
60%|██████ | 6/10 [00:01<00:01, 3.14it/s]/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/latest/lib/python3.9/site-packages/nilearn/masking.py:974: UserWarning: Data array used to create a new image contains 64-bit ints. This is likely due to creating the array with numpy and passing `int` as the `dtype`. Many tools such as FSL and SPM cannot deal with int64 in Nifti images, so for compatibility the data has been converted to int32.
return new_img_like(mask_img, unmasked, affine)
70%|███████ | 7/10 [00:02<00:00, 3.14it/s]/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/latest/lib/python3.9/site-packages/nilearn/masking.py:974: UserWarning: Data array used to create a new image contains 64-bit ints. This is likely due to creating the array with numpy and passing `int` as the `dtype`. Many tools such as FSL and SPM cannot deal with int64 in Nifti images, so for compatibility the data has been converted to int32.
return new_img_like(mask_img, unmasked, affine)
80%|████████ | 8/10 [00:02<00:00, 3.14it/s]/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/latest/lib/python3.9/site-packages/nilearn/masking.py:974: UserWarning: Data array used to create a new image contains 64-bit ints. This is likely due to creating the array with numpy and passing `int` as the `dtype`. Many tools such as FSL and SPM cannot deal with int64 in Nifti images, so for compatibility the data has been converted to int32.
return new_img_like(mask_img, unmasked, affine)
90%|█████████ | 9/10 [00:02<00:00, 3.14it/s]/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/latest/lib/python3.9/site-packages/nilearn/masking.py:974: UserWarning: Data array used to create a new image contains 64-bit ints. This is likely due to creating the array with numpy and passing `int` as the `dtype`. Many tools such as FSL and SPM cannot deal with int64 in Nifti images, so for compatibility the data has been converted to int32.
return new_img_like(mask_img, unmasked, affine)
100%|██████████| 10/10 [00:03<00:00, 3.13it/s]
100%|██████████| 10/10 [00:03<00:00, 3.11it/s]
Description:
('A kernel density (KDA) meta-analysis \\citep{wager2007meta} was performed '
'was performed with NiMARE 0.2.2+2.gbf7df33.dirty (RRID:SCR_017398; '
'\\citealt{Salo2023}), using a(n) KDA kernel. A KDA kernel '
'\\citep{wager2003valence,wager2004neuroimaging} was used to generate '
'study-wise modeled activation maps from coordinates. In this kernel method, '
'each coordinate is convolved with a sphere with a radius of 10.0 and a value '
"of 1. These spheres are then summed within each study to produce the study's "
'MA map. Summary statistics (OF values) were converted to p-values using an '
'approximate null distribution. The input dataset included 267 foci from 21 '
'experiments.')
References:
('@article{Salo2023,\n'
' doi = {10.52294/001c.87681},\n'
' url = {https://doi.org/10.52294/001c.87681},\n'
' year = {2023},\n'
' volume = {3},\n'
' pages = {1 - 32},\n'
' author = {Taylor Salo and Tal Yarkoni and Thomas E. Nichols and '
'Jean-Baptiste Poline and Murat Bilgel and Katherine L. Bottenhorn and Dorota '
'Jarecka and James D. Kent and Adam Kimbler and Dylan M. Nielson and Kendra '
'M. Oudyk and Julio A. Peraza and Alexandre Pérez and Puck C. Reeders and '
'Julio A. Yanes and Angela R. Laird},\n'
' title = {NiMARE: Neuroimaging Meta-Analysis Research Environment},\n'
' journal = {Aperture Neuro}\n'
'}\n'
'@article{wager2003valence,\n'
' title={Valence, gender, and lateralization of functional brain anatomy in '
'emotion: a meta-analysis of findings from neuroimaging},\n'
' author={Wager, Tor D and Phan, K Luan and Liberzon, Israel and Taylor, '
'Stephan F},\n'
' journal={Neuroimage},\n'
' volume={19},\n'
' number={3},\n'
' pages={513--531},\n'
' year={2003},\n'
' publisher={Elsevier},\n'
' url={https://doi.org/10.1016/S1053-8119(03)00078-8},\n'
' doi={10.1016/S1053-8119(03)00078-8}\n'
'}\n'
'@article{wager2004neuroimaging,\n'
' title={Neuroimaging studies of shifting attention: a meta-analysis},\n'
' author={Wager, Tor D and Jonides, John and Reading, Susan},\n'
' journal={Neuroimage},\n'
' volume={22},\n'
' number={4},\n'
' pages={1679--1693},\n'
' year={2004},\n'
' publisher={Elsevier},\n'
' url={https://doi.org/10.1016/j.neuroimage.2004.03.052},\n'
' doi={10.1016/j.neuroimage.2004.03.052}\n'
'}\n'
'@article{wager2007meta,\n'
' title={Meta-analysis of functional neuroimaging data: current and future '
'directions},\n'
' author={Wager, Tor D and Lindquist, Martin and Kaplan, Lauren},\n'
' journal={Social cognitive and affective neuroscience},\n'
' volume={2},\n'
' number={2},\n'
' pages={150--158},\n'
' year={2007},\n'
' publisher={Oxford University Press},\n'
' url={https://doi.org/10.1093/scan/nsm015},\n'
' doi={10.1093/scan/nsm015}\n'
'}')
Activation Likelihood Estimation
from nimare.meta.cbma.ale import ALE
meta = ALE()
results = meta.fit(dset)
corr = FWECorrector(method="montecarlo", n_iters=10, n_cores=1)
cres = corr.transform(results)
plot_stat_map(
results.get_map("z"),
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
threshold=0.1,
)
plot_stat_map(
cres.get_map("z_desc-size_level-cluster_corr-FWE_method-montecarlo"),
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
threshold=0.1,
)
print("Description:")
pprint(results.description_)
print("References:")
pprint(results.bibtex_)
0%| | 0/10 [00:00<?, ?it/s]
10%|█ | 1/10 [00:00<00:06, 1.40it/s]
20%|██ | 2/10 [00:01<00:05, 1.43it/s]
30%|███ | 3/10 [00:02<00:04, 1.44it/s]
40%|████ | 4/10 [00:02<00:04, 1.47it/s]
50%|█████ | 5/10 [00:03<00:03, 1.45it/s]
60%|██████ | 6/10 [00:04<00:02, 1.44it/s]
70%|███████ | 7/10 [00:04<00:02, 1.45it/s]
80%|████████ | 8/10 [00:05<00:01, 1.44it/s]
90%|█████████ | 9/10 [00:06<00:00, 1.44it/s]
100%|██████████| 10/10 [00:06<00:00, 1.45it/s]
100%|██████████| 10/10 [00:06<00:00, 1.45it/s]
Description:
('An activation likelihood estimation (ALE) meta-analysis '
'\\citep{turkeltaub2002meta,turkeltaub2012minimizing,eickhoff2012activation} '
'was performed with NiMARE 0.2.2+2.gbf7df33.dirty (RRID:SCR_017398; '
'\\citealt{Salo2023}), using a(n) ALE kernel. An ALE kernel '
'\\citep{eickhoff2012activation} was used to generate study-wise modeled '
'activation maps from coordinates. In this kernel method, each coordinate is '
'convolved with a Gaussian kernel with full-width at half max values '
'determined on a study-wise basis based on the study sample sizes according '
'to the formulae provided in \\cite{eickhoff2012activation}. For voxels with '
'overlapping kernels, the maximum value was retained. ALE values were '
'converted to p-values using an approximate null distribution '
'\\citep{eickhoff2012activation}. The input dataset included 267 foci from 21 '
'experiments, with a total of 334 participants.')
References:
('@article{Salo2023,\n'
' doi = {10.52294/001c.87681},\n'
' url = {https://doi.org/10.52294/001c.87681},\n'
' year = {2023},\n'
' volume = {3},\n'
' pages = {1 - 32},\n'
' author = {Taylor Salo and Tal Yarkoni and Thomas E. Nichols and '
'Jean-Baptiste Poline and Murat Bilgel and Katherine L. Bottenhorn and Dorota '
'Jarecka and James D. Kent and Adam Kimbler and Dylan M. Nielson and Kendra '
'M. Oudyk and Julio A. Peraza and Alexandre Pérez and Puck C. Reeders and '
'Julio A. Yanes and Angela R. Laird},\n'
' title = {NiMARE: Neuroimaging Meta-Analysis Research Environment},\n'
' journal = {Aperture Neuro}\n'
'}\n'
'@article{eickhoff2012activation,\n'
' title={Activation likelihood estimation meta-analysis revisited},\n'
' author={Eickhoff, Simon B and Bzdok, Danilo and Laird, Angela R and Kurth, '
'Florian and Fox, Peter T},\n'
' journal={Neuroimage},\n'
' volume={59},\n'
' number={3},\n'
' pages={2349--2361},\n'
' year={2012},\n'
' publisher={Elsevier},\n'
' url={https://doi.org/10.1016/j.neuroimage.2011.09.017},\n'
' doi={10.1016/j.neuroimage.2011.09.017}\n'
'}\n'
'@article{turkeltaub2002meta,\n'
' title={Meta-analysis of the functional neuroanatomy of single-word '
'reading: method and validation},\n'
' author={Turkeltaub, Peter E and Eden, Guinevere F and Jones, Karen M and '
'Zeffiro, Thomas A},\n'
' journal={Neuroimage},\n'
' volume={16},\n'
' number={3},\n'
' pages={765--780},\n'
' year={2002},\n'
' publisher={Elsevier},\n'
' url={https://doi.org/10.1006/nimg.2002.1131},\n'
' doi={10.1006/nimg.2002.1131}\n'
'}\n'
'@article{turkeltaub2012minimizing,\n'
' title={Minimizing within-experiment and within-group effects in activation '
'likelihood estimation meta-analyses},\n'
' author={Turkeltaub, Peter E and Eickhoff, Simon B and Laird, Angela R and '
'Fox, Mick and Wiener, Martin and Fox, Peter},\n'
' journal={Human brain mapping},\n'
' volume={33},\n'
' number={1},\n'
' pages={1--13},\n'
' year={2012},\n'
' publisher={Wiley Online Library},\n'
' url={https://doi.org/10.1002/hbm.21186},\n'
' doi={10.1002/hbm.21186}\n'
'}')
Specific Co-Activation Likelihood Estimation
Important
The SCALE algorithm is very memory intensive, so we don’t run it within the documentation.
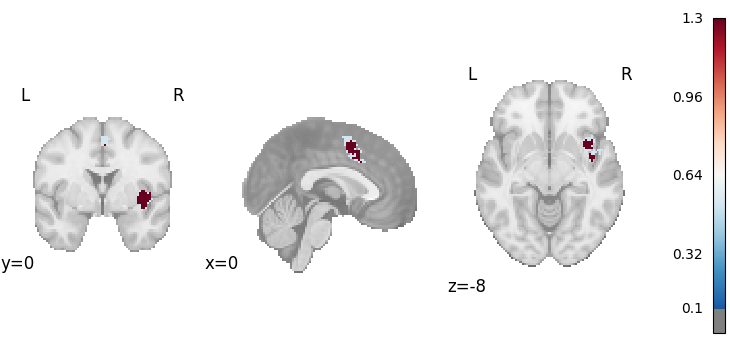
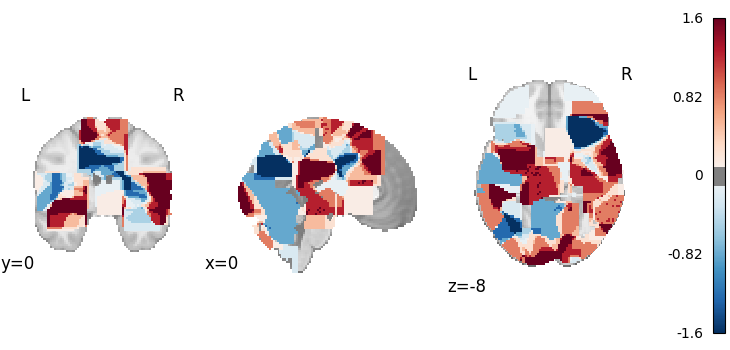
ALE-Based Subtraction Analysis
from nimare.meta.cbma.ale import ALESubtraction
meta = ALESubtraction(n_iters=10, n_cores=1)
results = meta.fit(dset1, dset2)
plot_stat_map(
results.get_map("z_desc-group1MinusGroup2"),
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
threshold=0.1,
)
print("Description:")
pprint(results.description_)
print("References:")
pprint(results.bibtex_)
0%| | 0/10 [00:00<?, ?it/s]
10%|█ | 1/10 [00:04<00:44, 4.97s/it]
20%|██ | 2/10 [00:05<00:17, 2.22s/it]
30%|███ | 3/10 [00:05<00:09, 1.34s/it]
40%|████ | 4/10 [00:05<00:05, 1.08it/s]
50%|█████ | 5/10 [00:06<00:03, 1.43it/s]
60%|██████ | 6/10 [00:06<00:02, 1.78it/s]
70%|███████ | 7/10 [00:06<00:01, 2.12it/s]
80%|████████ | 8/10 [00:07<00:00, 2.40it/s]
90%|█████████ | 9/10 [00:07<00:00, 2.65it/s]
100%|██████████| 10/10 [00:07<00:00, 2.85it/s]
100%|██████████| 10/10 [00:07<00:00, 1.32it/s]
0%| | 0/228483 [00:00<?, ?it/s]
0%| | 2/228483 [00:00<15:13, 250.02it/s]
Description:
('An activation likelihood estimation (ALE) subtraction analysis '
'\\citep{laird2005ale,eickhoff2012activation} was performed with NiMARE '
'v0.2.2+2.gbf7df33.dirty (RRID:SCR_017398; \\citealt{Salo2023}), using a(n) '
'ALE kernel. An ALE kernel \\citep{eickhoff2012activation} was used to '
'generate study-wise modeled activation maps from coordinates. In this kernel '
'method, each coordinate is convolved with a Gaussian kernel with full-width '
'at half max values determined on a study-wise basis based on the study '
'sample sizes according to the formulae provided in '
'\\cite{eickhoff2012activation}. For voxels with overlapping kernels, the '
'maximum value was retained. The subtraction analysis was implemented '
"according to NiMARE's \\citep{Salo2023} approach, which differs from the "
'original version. In this version, ALE-difference scores are calculated '
'between the two datasets, for all voxels in the mask, rather than for voxels '
'significant in the main effects analyses of the two datasets. Next, '
'voxel-wise null distributions of ALE-difference scores were generated via a '
'randomized group assignment procedure, in which the studies in the two '
'datasets were randomly reassigned and ALE-difference scores were calculated '
'for the randomized datasets. This randomization procedure was repeated 10 '
'times to build the null distributions. The significance of the original '
'ALE-difference scores was assessed using a two-sided statistical test. The '
'null distributions were assumed to be asymmetric, as ALE-difference scores '
'will be skewed based on the sample sizes of the two datasets. The first '
'input dataset (group1) included 147 foci from 10 experiments, with a total '
'of 153 participants. The second input dataset (group2) included 120 foci '
'from 11 experiments, with a total of 181 participants. ')
References:
('@article{Salo2023,\n'
' doi = {10.52294/001c.87681},\n'
' url = {https://doi.org/10.52294/001c.87681},\n'
' year = {2023},\n'
' volume = {3},\n'
' pages = {1 - 32},\n'
' author = {Taylor Salo and Tal Yarkoni and Thomas E. Nichols and '
'Jean-Baptiste Poline and Murat Bilgel and Katherine L. Bottenhorn and Dorota '
'Jarecka and James D. Kent and Adam Kimbler and Dylan M. Nielson and Kendra '
'M. Oudyk and Julio A. Peraza and Alexandre Pérez and Puck C. Reeders and '
'Julio A. Yanes and Angela R. Laird},\n'
' title = {NiMARE: Neuroimaging Meta-Analysis Research Environment},\n'
' journal = {Aperture Neuro}\n'
'}\n'
'@article{eickhoff2012activation,\n'
' title={Activation likelihood estimation meta-analysis revisited},\n'
' author={Eickhoff, Simon B and Bzdok, Danilo and Laird, Angela R and Kurth, '
'Florian and Fox, Peter T},\n'
' journal={Neuroimage},\n'
' volume={59},\n'
' number={3},\n'
' pages={2349--2361},\n'
' year={2012},\n'
' publisher={Elsevier},\n'
' url={https://doi.org/10.1016/j.neuroimage.2011.09.017},\n'
' doi={10.1016/j.neuroimage.2011.09.017}\n'
'}\n'
'@article{laird2005ale,\n'
' title={ALE meta-analysis: Controlling the false discovery rate and '
'performing statistical contrasts},\n'
' author={Laird, Angela R and Fox, P Mickle and Price, Cathy J and Glahn, '
'David C and Uecker, Angela M and Lancaster, Jack L and Turkeltaub, Peter E '
'and Kochunov, Peter and Fox, Peter T},\n'
' journal={Human brain mapping},\n'
' volume={25},\n'
' number={1},\n'
' pages={155--164},\n'
' year={2005},\n'
' publisher={Wiley Online Library},\n'
' url={https://doi.org/10.1002/hbm.20136},\n'
' doi={10.1002/hbm.20136}\n'
'}')
Total running time of the script: (1 minutes 16.278 seconds)