Note
Go to the end to download the full example code
KernelTransformers and CBMA
KernelTransformer classes are tools for converting individual studies’
coordinates into images.
For coordinate-based meta-analyses, individual studies’ statistical maps are mimicked by generating “modeled activation” (MA) maps from the coordinates. These MA maps are used in the CBMA algorithms, although the specific method used to generate the MA maps differs by algorithm.
This example provides an introduction to the KernelTransformer class and
a tour of available types.
import os
import matplotlib.pyplot as plt
from nilearn.plotting import plot_stat_map
Load Dataset
from nimare.dataset import Dataset
from nimare.utils import get_resource_path
dset_file = os.path.join(get_resource_path(), "nidm_pain_dset.json")
dset = Dataset(dset_file)
# First, let us reduce this Dataset to only two studies
dset = dset.slice(dset.ids[2:4])
Kernels ingest Datasets and can produce a few types of outputs
from nimare.meta.kernel import MKDAKernel
# First, the kernel should be initialized with any parameters.
kernel = MKDAKernel()
# Then, the ``transform`` method takes in the Dataset and produces the MA maps.
output = kernel.transform(dset)
return_type="image" returns a list of 3D niimg objects.
This is the default option.
image_output = kernel.transform(dset, return_type="image")
print(type(image_output))
print(type(image_output[0]))
print(image_output[0].shape)
<class 'list'>
<class 'nibabel.nifti1.Nifti1Image'>
(91, 109, 91)
return_type="array" returns a 2D numpy array
array_output = kernel.transform(dset, return_type="array")
print(type(array_output))
print(array_output.shape)
<class 'numpy.ndarray'>
(2, 228483)
There is also an option to return an updated Dataset
(return_type="dataset"), with the MA maps saved as nifti files and
references in the Dataset’s images attribute.
However, this will only work if the Dataset has a location set for its
images.
try:
dataset_output = kernel.transform(dset, return_type="dataset")
except ValueError as error:
print(error)
Argument "return_type" must be "image", "array", "summary_array", "sparse".
Each kernel can accept certain parameters that control behavior
You can see what options are available via the API documentation or through the help string.
help(MKDAKernel)
Help on class MKDAKernel in module nimare.meta.kernel:
class MKDAKernel(KDAKernel)
| MKDAKernel(r=10, value=1, memory=Memory(location=None), memory_level=0)
|
| Generate MKDA modeled activation images from coordinates.
|
| .. versionchanged:: 0.2.1
|
| - New parameters: ``memory`` and ``memory_level`` for memory caching.
| - Add new parameter ``return_type`` to transform method.
|
| .. versionchanged:: 0.0.13
|
| - Remove "dataset" `return_type` option.
|
| .. versionchanged:: 0.0.12
|
| * Remove low-memory option in favor of sparse arrays for kernel transformers.
|
| Parameters
| ----------
| r : :obj:`int`, default=10
| Sphere radius, in mm.
| value : :obj:`int`, default=1
| Value for sphere.
| memory : instance of :class:`joblib.Memory`, :obj:`str`, or :class:`pathlib.Path`
| Used to cache the output of a function. By default, no caching is done.
| If a :obj:`str` is given, it is the path to the caching directory.
| memory_level : :obj:`int`, default=0
| Rough estimator of the amount of memory used by caching.
| Higher value means more memory for caching. Zero means no caching.
|
| Method resolution order:
| MKDAKernel
| KDAKernel
| KernelTransformer
| nimare.base.NiMAREBase
| nilearn._utils.cache_mixin.CacheMixin
| builtins.object
|
| Data and other attributes defined here:
|
| __abstractmethods__ = frozenset()
|
| __slotnames__ = []
|
| ----------------------------------------------------------------------
| Methods inherited from KDAKernel:
|
| __init__(self, r=10, value=1, memory=Memory(location=None), memory_level=0)
| Initialize self. See help(type(self)) for accurate signature.
|
| ----------------------------------------------------------------------
| Methods inherited from KernelTransformer:
|
| transform(self, dataset, masker=None, return_type='image')
| Generate modeled activation images for each Contrast in dataset.
|
| Parameters
| ----------
| dataset : :obj:`~nimare.dataset.Dataset` or :obj:`pandas.DataFrame`
| Dataset for which to make images. Can be a DataFrame if necessary.
| masker : img_like or None, optional
| Mask to apply to MA maps. Required if ``dataset`` is a DataFrame.
| If None (and ``dataset`` is a Dataset), the Dataset's masker attribute will be used.
| Default is None.
| return_type : {'sparse', 'array', 'image', 'summary_array'}, optional
| Whether to return a sparse matrix ('sparse'), a numpy array ('array'),
| or a list of niimgs ('image').
| Default is 'image'.
|
| Returns
| -------
| imgs : (C x V) :class:`numpy.ndarray` or :obj:`list` of :class:`nibabel.Nifti1Image` or :class:`~nimare.dataset.Dataset`
| If return_type is 'sparse', a 4D sparse array (E x S), where E is
| the number of unique experiments, and the remaining 3 dimensions are
| equal to `shape` of the images.
| If return_type is 'array', a 2D numpy array (C x V), where C is
| contrast and V is voxel.
| If return_type is 'summary_array', a 1D numpy array (V,) containing
| a summary measure for each voxel that has been combined across experiments.
| If return_type is 'image', a list of modeled activation images
| (one for each of the Contrasts in the input dataset).
|
| Attributes
| ----------
| filename_pattern : str
| Filename pattern for MA maps. If :meth:`_infer_names` is executed.
| image_type : str
| Name of the corresponding column in the Dataset.images DataFrame.
| If :meth:`_infer_names` is executed.
|
| ----------------------------------------------------------------------
| Methods inherited from nimare.base.NiMAREBase:
|
| __repr__(self)
| Show basic NiMARE class representation.
|
| Specifically, this shows the name of the class, along with any parameters
| that are **not** set to the default.
|
| get_params(self, deep=True)
| Get parameters for this estimator.
|
| Parameters
| ----------
| deep : :obj:`bool`, default=True
| If True, will return the parameters for this estimator and
| contained subobjects that are estimators.
|
| Returns
| -------
| params : :obj:`dict`
| Parameter names mapped to their values.
|
| save(self, filename, compress=True)
| Pickle the class instance to the provided file.
|
| Parameters
| ----------
| filename : :obj:`str`
| File to which object will be saved.
| compress : :obj:`bool`, optional
| If True, the file will be compressed with gzip. Otherwise, the
| uncompressed version will be saved. Default = True.
|
| set_params(self, **params)
| Set the parameters of this estimator.
|
| The method works on simple estimators as well as on nested objects
| (such as pipelines). The latter have parameters of the form
| ``<component>__<parameter>`` so that it's possible to update each
| component of a nested object.
|
| Returns
| -------
| self
|
| ----------------------------------------------------------------------
| Class methods inherited from nimare.base.NiMAREBase:
|
| load(filename, compressed=True) from abc.ABCMeta
| Load a pickled class instance from file.
|
| Parameters
| ----------
| filename : :obj:`str`
| Name of file containing object.
| compressed : :obj:`bool`, default=True
| If True, the file is assumed to be compressed and gzip will be used
| to load it. Otherwise, it will assume that the file is not
| compressed. Default = True.
|
| Returns
| -------
| obj : class object
| Loaded class object.
|
| ----------------------------------------------------------------------
| Data descriptors inherited from nilearn._utils.cache_mixin.CacheMixin:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
For example, MKDAKernel kernel accepts an r
argument to control the radius of the kernel.
RADIUS_VALUES = [4, 8, 12]
fig, axes = plt.subplots(ncols=3, figsize=(20, 10))
for i, radius in enumerate(RADIUS_VALUES):
kernel = MKDAKernel(r=radius)
ma_maps = kernel.transform(dset, return_type="image")
plot_stat_map(
ma_maps[0],
display_mode="z",
cut_coords=[-2],
title=f"r={radius}mm",
axes=axes[i],
draw_cross=False,
annotate=False,
colorbar=False,
cmap="RdBu_r",
)
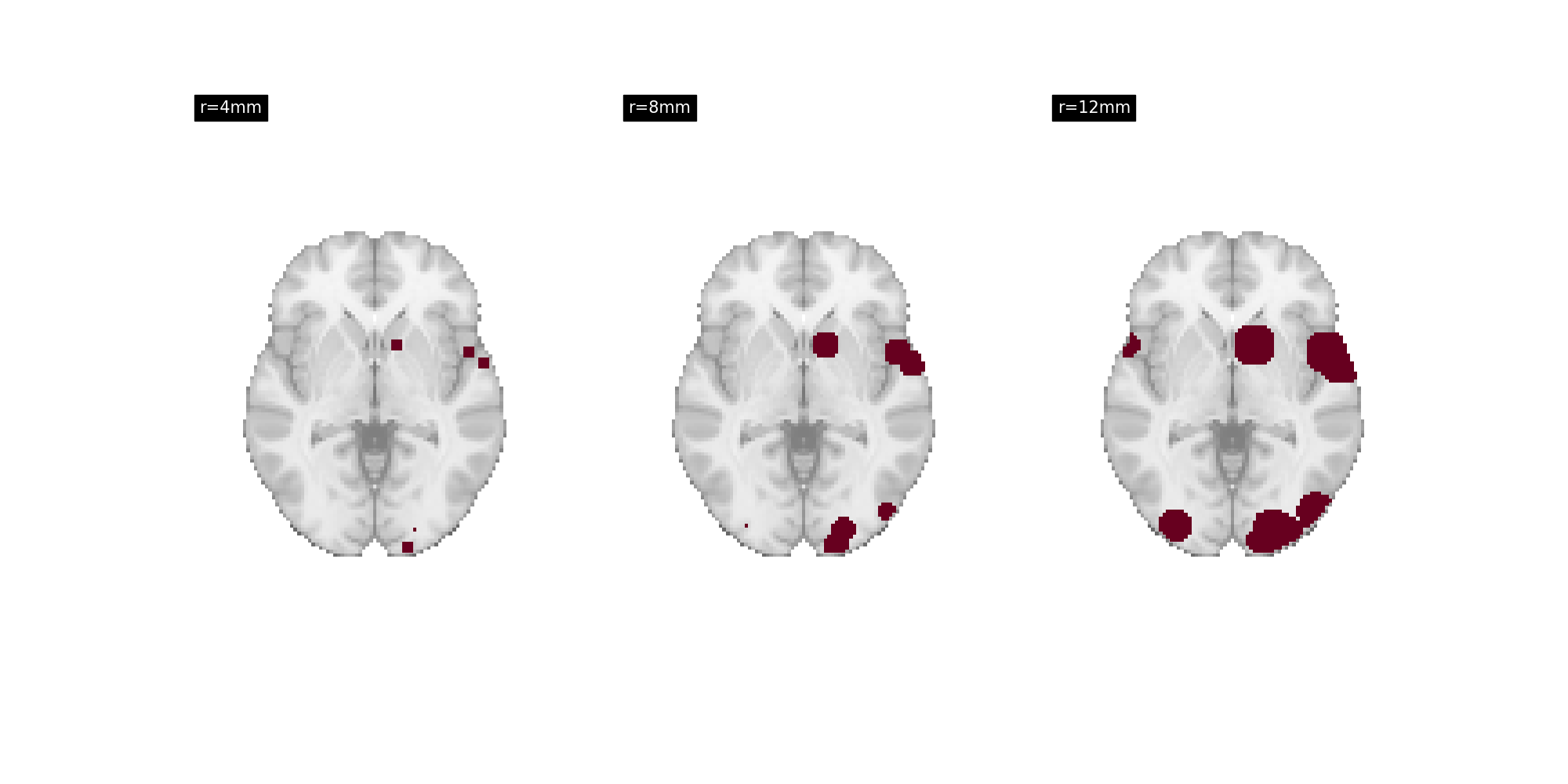
There are several kernels available
MKDAKernel convolves coordinates with a
sphere and takes the union across voxels.
kernel = MKDAKernel(r=10)
ma_maps = kernel.transform(dset, return_type="image")
plot_stat_map(
ma_maps[0],
cut_coords=[-2, -10, -4],
title="MKDA",
draw_cross=False,
cmap="RdBu_r",
)
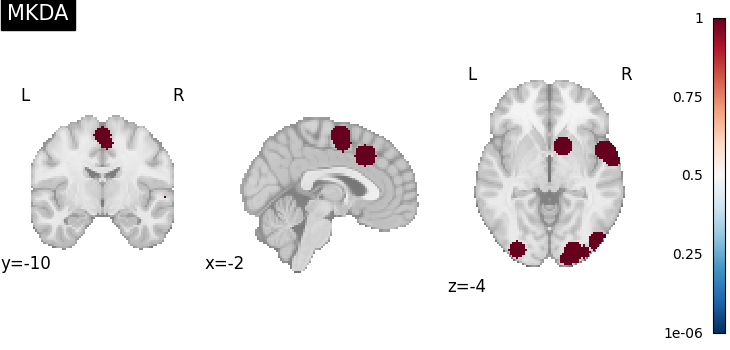
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7fe9231feb20>
KDAKernel convolves coordinates with a
sphere as well, but takes the sum across voxels.
from nimare.meta.kernel import KDAKernel
kernel = KDAKernel(r=10)
ma_maps = kernel.transform(dset, return_type="image")
plot_stat_map(
ma_maps[0],
cut_coords=[-2, -10, -4],
title="KDA",
draw_cross=False,
cmap="RdBu_r",
)
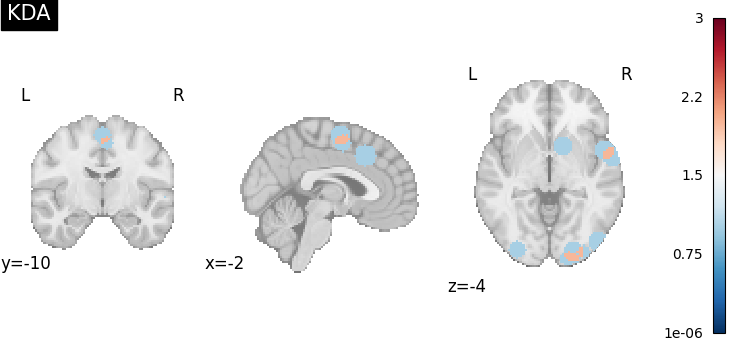
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7fe93dea0fa0>
ALEKernel convolves coordinates with a 3D
Gaussian, for which the FWHM is determined by the sample size of each study.
This sample size will be inferred automatically, if that information is
available in the Dataset, or it can be set as a constant value across all
studies in the Dataset with the sample_size argument.
from nimare.meta.kernel import ALEKernel
kernel = ALEKernel(sample_size=20)
ma_maps = kernel.transform(dset, return_type="image")
plot_stat_map(
ma_maps[0],
cut_coords=[-2, -10, -4],
title="ALE",
draw_cross=False,
cmap="RdBu_r",
)
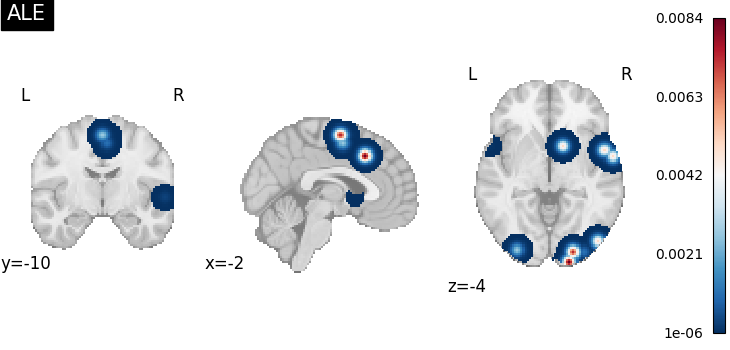
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7fe93da3aa00>
Total running time of the script: (0 minutes 15.288 seconds)