Note
Go to the end to download the full example code
Use NeuroVault statistical maps in NiMARE
Download statistical maps from NeuroVault, then use them in a meta-analysis, with NiMARE.
import matplotlib.pyplot as plt
from nilearn.plotting import plot_stat_map
Conversion of Statistical Maps
Some of the statistical maps are T statistics and others are Z statistics.
To perform a Fisher’s meta analysis, we need all Z maps.
Thoughtfully, NiMARE has a class named ImageTransformer that will
help us.
from nimare.transforms import ImageTransformer
# Not all studies have Z maps!
dset.images[["z"]]
z_transformer = ImageTransformer(target="z")
dset = z_transformer.transform(dset)
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/image/resampling.py:663: RuntimeWarning: NaNs or infinite values are present in the data passed to resample. This is a bad thing as they make resampling ill-defined and much slower.
_resample_one_img(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
All studies now have Z maps!
dset.images[["z"]]
Run a Meta-Analysis
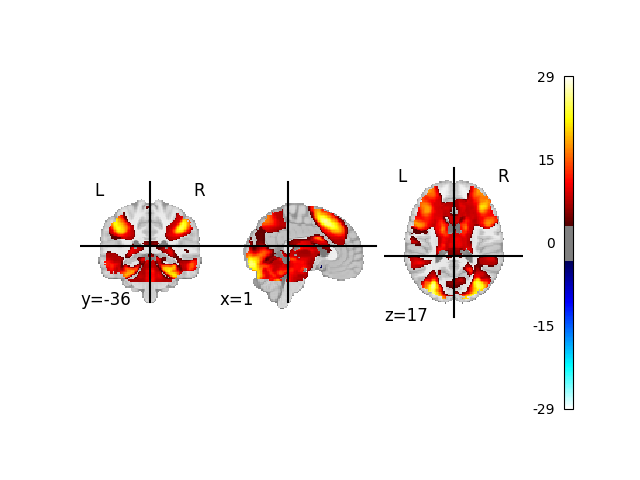
With the missing Z maps filled in, we can run a Meta-Analysis and plot our results
from nimare.meta.ibma import Fishers
# The default template has a slightly different, but completely compatible,
# affine than the NeuroVault images, so we allow the Estimator to resample
# images during the fitting process.
meta = Fishers(resample=True)
meta_res = meta.fit(dset)
fig, ax = plt.subplots()
display = plot_stat_map(meta_res.get_map("z"), threshold=3.3, axes=ax, figure=fig)
fig.show()
# The result may look questionable, but this code provides
# a template on how to use neurovault in your meta analysis.
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/numpy/core/numeric.py:330: RuntimeWarning: invalid value encountered in cast
multiarray.copyto(a, fill_value, casting='unsafe')
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/_utils/niimg.py:61: UserWarning: Non-finite values detected. These values will be replaced with zeros.
warn(
Total running time of the script: (0 minutes 18.575 seconds)