Note
Go to the end to download the full example code
Image-based meta-analysis algorithms
A tour of IBMA algorithms in NiMARE.
This tutorial is intended to provide a brief description and example of each of the IBMA algorithms implemented in NiMARE. For a more detailed introduction to the elements of an image-based meta-analysis, see other stuff.
from nilearn.plotting import plot_stat_map
Download data
Note
The data used in this example come from a collection of NIDM-Results packs downloaded from Neurovault collection 1425, uploaded by Dr. Camille Maumet.
from nimare.extract import download_nidm_pain
dset_dir = download_nidm_pain()
Load Dataset
import os
from pprint import pprint
from nimare.dataset import Dataset
from nimare.transforms import ImageTransformer
from nimare.utils import get_resource_path
dset_file = os.path.join(get_resource_path(), "nidm_pain_dset.json")
dset = Dataset(dset_file)
dset.update_path(dset_dir)
# Calculate missing images
xformer = ImageTransformer(target=["varcope", "z"])
dset = xformer.transform(dset)
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
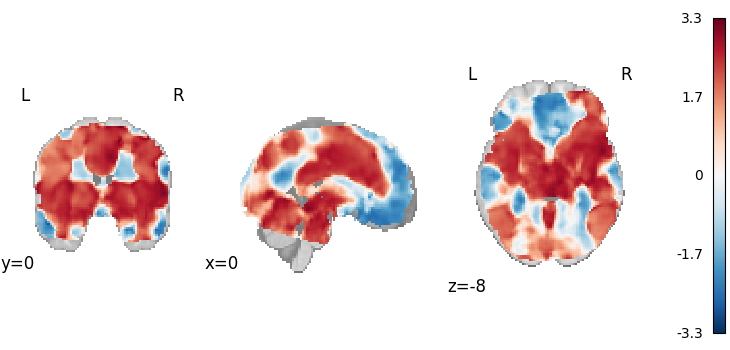
Stouffer’s
from nimare.meta.ibma import Stouffers
meta = Stouffers(use_sample_size=False)
results = meta.fit(dset)
plot_stat_map(
results.get_map("z"),
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
)
print("Description:")
pprint(results.description_)
print("References:")
pprint(results.bibtex_)
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/_utils/niimg.py:61: UserWarning: Non-finite values detected. These values will be replaced with zeros.
warn(
Description:
('An image-based meta-analysis was performed with NiMARE '
'0.2.2+0.g07ac3b6.dirty (RRID:SCR_017398; \\citealt{Salo2023}) on 21 '
'z-statistic images using the Stouffer method \\citep{stouffer1949american}.')
References:
('@article{Salo2023,\n'
' doi = {10.52294/001c.87681},\n'
' url = {https://doi.org/10.52294/001c.87681},\n'
' year = {2023},\n'
' volume = {3},\n'
' pages = {1 - 32},\n'
' author = {Taylor Salo and Tal Yarkoni and Thomas E. Nichols and '
'Jean-Baptiste Poline and Murat Bilgel and Katherine L. Bottenhorn and Dorota '
'Jarecka and James D. Kent and Adam Kimbler and Dylan M. Nielson and Kendra '
'M. Oudyk and Julio A. Peraza and Alexandre Pérez and Puck C. Reeders and '
'Julio A. Yanes and Angela R. Laird},\n'
' title = {NiMARE: Neuroimaging Meta-Analysis Research Environment},\n'
' journal = {Aperture Neuro}\n'
'}\n'
'@article{stouffer1949american,\n'
' title={The american soldier: Adjustment during army life.(studies in '
'social psychology in world war ii), vol. 1},\n'
' author={Stouffer, Samuel A and Suchman, Edward A and DeVinney, Leland C '
'and Star, Shirley A and Williams Jr, Robin M},\n'
' journal={Studies in social psychology in World War II},\n'
' year={1949},\n'
' publisher={Princeton Univ. Press}\n'
'}')
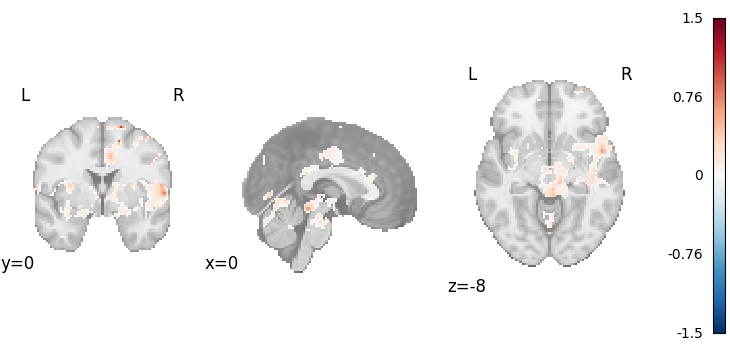
Stouffer’s with weighting by sample size
meta = Stouffers(use_sample_size=True)
results = meta.fit(dset)
plot_stat_map(
results.get_map("z"),
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
)
print("Description:")
pprint(results.description_)
print("References:")
pprint(results.bibtex_)
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/_utils/niimg.py:61: UserWarning: Non-finite values detected. These values will be replaced with zeros.
warn(
Description:
('An image-based meta-analysis was performed with NiMARE '
'0.2.2+0.g07ac3b6.dirty (RRID:SCR_017398; \\citealt{Salo2023}) on 21 '
'z-statistic images using the Stouffer method \\citep{stouffer1949american}, '
'with studies weighted by the square root of the study sample sizes, per '
'\\cite{zaykin2011optimally}.')
References:
('@article{Salo2023,\n'
' doi = {10.52294/001c.87681},\n'
' url = {https://doi.org/10.52294/001c.87681},\n'
' year = {2023},\n'
' volume = {3},\n'
' pages = {1 - 32},\n'
' author = {Taylor Salo and Tal Yarkoni and Thomas E. Nichols and '
'Jean-Baptiste Poline and Murat Bilgel and Katherine L. Bottenhorn and Dorota '
'Jarecka and James D. Kent and Adam Kimbler and Dylan M. Nielson and Kendra '
'M. Oudyk and Julio A. Peraza and Alexandre Pérez and Puck C. Reeders and '
'Julio A. Yanes and Angela R. Laird},\n'
' title = {NiMARE: Neuroimaging Meta-Analysis Research Environment},\n'
' journal = {Aperture Neuro}\n'
'}\n'
'@article{stouffer1949american,\n'
' title={The american soldier: Adjustment during army life.(studies in '
'social psychology in world war ii), vol. 1},\n'
' author={Stouffer, Samuel A and Suchman, Edward A and DeVinney, Leland C '
'and Star, Shirley A and Williams Jr, Robin M},\n'
' journal={Studies in social psychology in World War II},\n'
' year={1949},\n'
' publisher={Princeton Univ. Press}\n'
'}\n'
'@article{zaykin2011optimally,\n'
' title={Optimally weighted Z-test is a powerful method for combining '
'probabilities in meta-analysis},\n'
' author={Zaykin, Dmitri V},\n'
' journal={Journal of evolutionary biology},\n'
' volume={24},\n'
' number={8},\n'
' pages={1836--1841},\n'
' year={2011},\n'
' publisher={Wiley Online Library},\n'
' url={https://doi.org/10.1111/j.1420-9101.2011.02297.x},\n'
' doi={10.1111/j.1420-9101.2011.02297.x}\n'
'}')
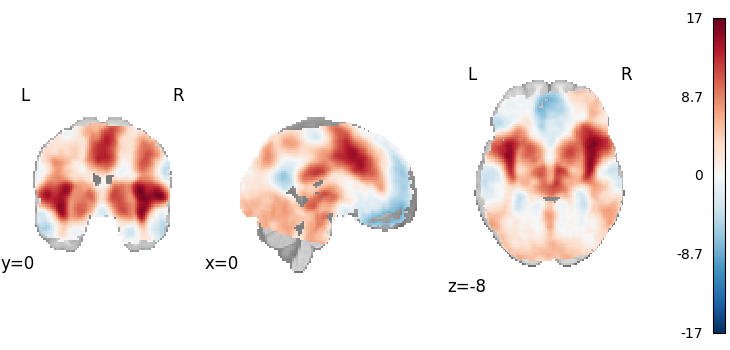
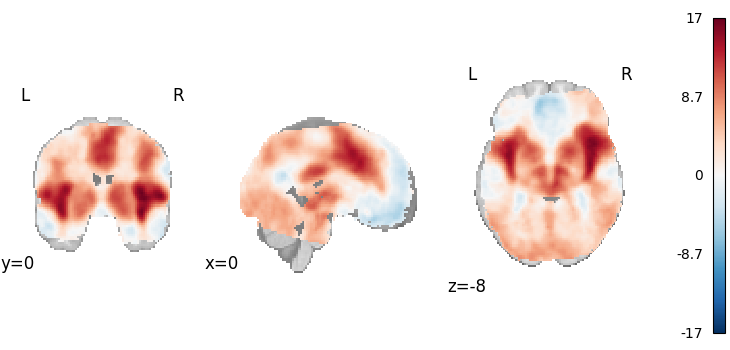
Fisher’s
from nimare.meta.ibma import Fishers
meta = Fishers()
results = meta.fit(dset)
plot_stat_map(
results.get_map("z"),
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
)
print("Description:")
pprint(results.description_)
print("References:")
pprint(results.bibtex_)
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/_utils/niimg.py:61: UserWarning: Non-finite values detected. These values will be replaced with zeros.
warn(
Description:
('An image-based meta-analysis was performed with NiMARE '
'0.2.2+0.g07ac3b6.dirty (RRID:SCR_017398; \\citealt{Salo2023}) on 21 '
'z-statistic images using the Fisher combined probability method '
'\\citep{fisher1946statistical}.')
References:
('@article{Salo2023,\n'
' doi = {10.52294/001c.87681},\n'
' url = {https://doi.org/10.52294/001c.87681},\n'
' year = {2023},\n'
' volume = {3},\n'
' pages = {1 - 32},\n'
' author = {Taylor Salo and Tal Yarkoni and Thomas E. Nichols and '
'Jean-Baptiste Poline and Murat Bilgel and Katherine L. Bottenhorn and Dorota '
'Jarecka and James D. Kent and Adam Kimbler and Dylan M. Nielson and Kendra '
'M. Oudyk and Julio A. Peraza and Alexandre Pérez and Puck C. Reeders and '
'Julio A. Yanes and Angela R. Laird},\n'
' title = {NiMARE: Neuroimaging Meta-Analysis Research Environment},\n'
' journal = {Aperture Neuro}\n'
'}\n'
'@article{fisher1946statistical,\n'
' title={Statistical methods for research workers.},\n'
' author={Fisher, Ronald Aylmer and others},\n'
' journal={Statistical methods for research workers.},\n'
' number={10th. ed.},\n'
' year={1946},\n'
' publisher={Oliver and Boyd}\n'
'}')
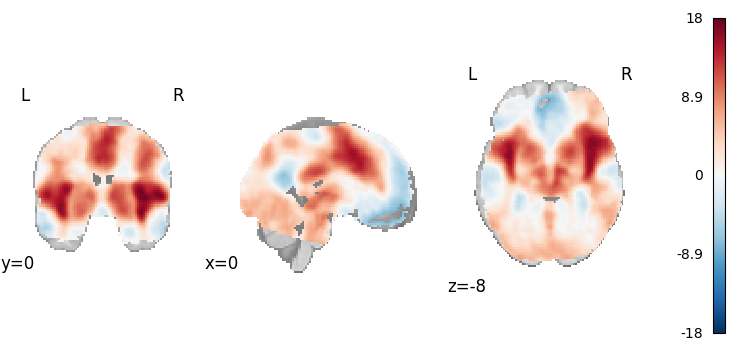
Permuted OLS
from nimare.correct import FWECorrector
from nimare.meta.ibma import PermutedOLS
meta = PermutedOLS(two_sided=True)
results = meta.fit(dset)
plot_stat_map(
results.get_map("z"),
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
)
corrector = FWECorrector(method="montecarlo", n_iters=100, n_cores=1)
cresult = corrector.transform(results)
plot_stat_map(
cresult.get_map("z_level-voxel_corr-FWE_method-montecarlo"),
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
)
print("Description:")
pprint(cresult.description_)
print("References:")
pprint(cresult.bibtex_)
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/_utils/niimg.py:61: UserWarning: Non-finite values detected. These values will be replaced with zeros.
warn(
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/_utils/niimg.py:61: UserWarning: Non-finite values detected. These values will be replaced with zeros.
warn(
Description:
('An image-based meta-analysis was performed with NiMARE '
'0.2.2+0.g07ac3b6.dirty (RRID:SCR_017398; \\citealt{Salo2023}), on 21 beta '
"images using Nilearn's \\citep{10.3389/fninf.2014.00014} permuted ordinary "
'least squares method. Family-wise error rate correction was performed using '
"Nilearn's \\citep{10.3389/fninf.2014.00014} permuted OLS method, in which "
'null distributions of test statistics were estimated using the max-value '
'permutation method detailed in \\cite{freedman1983nonstochastic}. 100 '
'iterations were performed to generate the null distribution.')
References:
('@article{10.3389/fninf.2014.00014,\n'
' title={Machine learning for neuroimaging with scikit-learn},\n'
' author={Abraham, Alexandre and Pedregosa, Fabian and Eickenberg, Michael '
'and Gervais, Philippe and Mueller, Andreas and Kossaifi, Jean and Gramfort, '
'Alexandre and Thirion, Bertrand and Varoquaux, Gael},\n'
'\tjournal={Frontiers in Neuroinformatics},\n'
'\tvolume={8},\n'
'\tyear={2014},\n'
'\turl={https://www.frontiersin.org/article/10.3389/fninf.2014.00014},\n'
'\tdoi={10.3389/fninf.2014.00014},\n'
'\tissn={1662-5196}\n'
'}\n'
'@article{Salo2023,\n'
' doi = {10.52294/001c.87681},\n'
' url = {https://doi.org/10.52294/001c.87681},\n'
' year = {2023},\n'
' volume = {3},\n'
' pages = {1 - 32},\n'
' author = {Taylor Salo and Tal Yarkoni and Thomas E. Nichols and '
'Jean-Baptiste Poline and Murat Bilgel and Katherine L. Bottenhorn and Dorota '
'Jarecka and James D. Kent and Adam Kimbler and Dylan M. Nielson and Kendra '
'M. Oudyk and Julio A. Peraza and Alexandre Pérez and Puck C. Reeders and '
'Julio A. Yanes and Angela R. Laird},\n'
' title = {NiMARE: Neuroimaging Meta-Analysis Research Environment},\n'
' journal = {Aperture Neuro}\n'
'}\n'
'@article{freedman1983nonstochastic,\n'
' title={A nonstochastic interpretation of reported significance levels},\n'
' author={Freedman, David and Lane, David},\n'
' journal={Journal of Business \\& Economic Statistics},\n'
' volume={1},\n'
' number={4},\n'
' pages={292--298},\n'
' year={1983},\n'
' publisher={Taylor \\& Francis}\n'
'}')
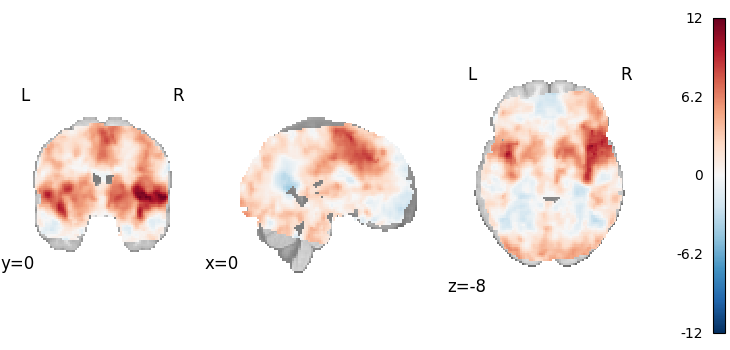
Weighted Least Squares
from nimare.meta.ibma import WeightedLeastSquares
meta = WeightedLeastSquares(tau2=0)
results = meta.fit(dset)
plot_stat_map(
results.get_map("z"),
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
)
print("Description:")
pprint(results.description_)
print("References:")
pprint(results.bibtex_)
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/_utils/niimg.py:61: UserWarning: Non-finite values detected. These values will be replaced with zeros.
warn(
Description:
('An image-based meta-analysis was performed with NiMARE '
'0.2.2+0.g07ac3b6.dirty (RRID:SCR_017398; \\citealt{Salo2023}), on 21 beta '
'images using the Weighted Least Squares approach '
'\\citep{brockwell2001comparison}, with an a priori tau-squared value of 0 '
'defined across all voxels.')
References:
('@article{Salo2023,\n'
' doi = {10.52294/001c.87681},\n'
' url = {https://doi.org/10.52294/001c.87681},\n'
' year = {2023},\n'
' volume = {3},\n'
' pages = {1 - 32},\n'
' author = {Taylor Salo and Tal Yarkoni and Thomas E. Nichols and '
'Jean-Baptiste Poline and Murat Bilgel and Katherine L. Bottenhorn and Dorota '
'Jarecka and James D. Kent and Adam Kimbler and Dylan M. Nielson and Kendra '
'M. Oudyk and Julio A. Peraza and Alexandre Pérez and Puck C. Reeders and '
'Julio A. Yanes and Angela R. Laird},\n'
' title = {NiMARE: Neuroimaging Meta-Analysis Research Environment},\n'
' journal = {Aperture Neuro}\n'
'}\n'
'@article{brockwell2001comparison,\n'
' title={A comparison of statistical methods for meta-analysis},\n'
' author={Brockwell, Sarah E and Gordon, Ian R},\n'
' journal={Statistics in medicine},\n'
' volume={20},\n'
' number={6},\n'
' pages={825--840},\n'
' year={2001},\n'
' publisher={Wiley Online Library},\n'
' url={https://doi.org/10.1002/sim.650},\n'
' doi={10.1002/sim.650}\n'
'}')
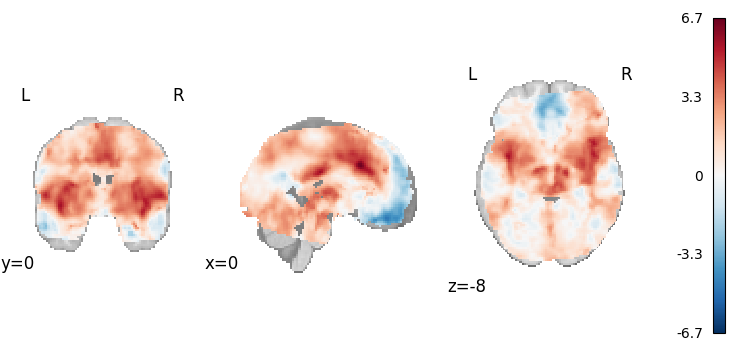
DerSimonian-Laird
from nimare.meta.ibma import DerSimonianLaird
meta = DerSimonianLaird()
results = meta.fit(dset)
plot_stat_map(
results.get_map("z"),
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
)
print("Description:")
pprint(results.description_)
print("References:")
pprint(results.bibtex_)
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/_utils/niimg.py:61: UserWarning: Non-finite values detected. These values will be replaced with zeros.
warn(
Description:
('An image-based meta-analysis was performed with NiMARE '
'0.2.2+0.g07ac3b6.dirty (RRID:SCR_017398; \\citealt{Salo2023}), on 21 beta '
'and variance images using the DerSimonian-Laird method '
'\\citep{dersimonian1986meta}, in which tau-squared is estimated on a '
'voxel-wise basis using the method-of-moments approach '
'\\citep{dersimonian1986meta,kosmidis2017improving}.')
References:
('@article{Salo2023,\n'
' doi = {10.52294/001c.87681},\n'
' url = {https://doi.org/10.52294/001c.87681},\n'
' year = {2023},\n'
' volume = {3},\n'
' pages = {1 - 32},\n'
' author = {Taylor Salo and Tal Yarkoni and Thomas E. Nichols and '
'Jean-Baptiste Poline and Murat Bilgel and Katherine L. Bottenhorn and Dorota '
'Jarecka and James D. Kent and Adam Kimbler and Dylan M. Nielson and Kendra '
'M. Oudyk and Julio A. Peraza and Alexandre Pérez and Puck C. Reeders and '
'Julio A. Yanes and Angela R. Laird},\n'
' title = {NiMARE: Neuroimaging Meta-Analysis Research Environment},\n'
' journal = {Aperture Neuro}\n'
'}\n'
'@article{dersimonian1986meta,\n'
' title={Meta-analysis in clinical trials},\n'
' author={DerSimonian, Rebecca and Laird, Nan},\n'
' journal={Controlled clinical trials},\n'
' volume={7},\n'
' number={3},\n'
' pages={177--188},\n'
' year={1986},\n'
' publisher={Elsevier}\n'
'}\n'
'@article{kosmidis2017improving,\n'
' title={Improving the accuracy of likelihood-based inference in '
'meta-analysis and meta-regression},\n'
' author={Kosmidis, Ioannis and Guolo, Annamaria and Varin, Cristiano},\n'
' journal={Biometrika},\n'
' volume={104},\n'
' number={2},\n'
' pages={489--496},\n'
' year={2017},\n'
' publisher={Oxford University Press},\n'
' url={https://doi.org/10.1093/biomet/asx001},\n'
' doi={10.1093/biomet/asx001}\n'
'}')
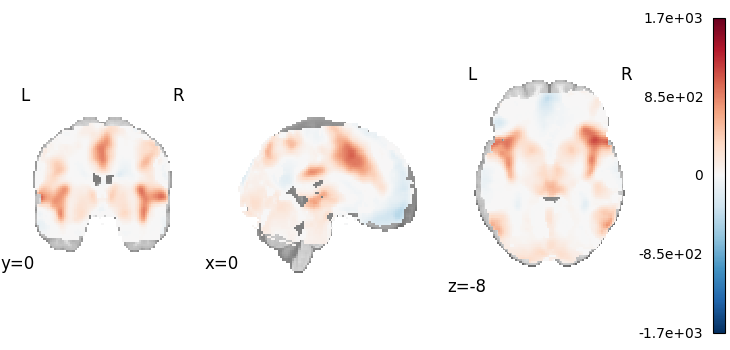
Hedges
from nimare.meta.ibma import Hedges
meta = Hedges()
results = meta.fit(dset)
plot_stat_map(
results.get_map("z"),
cut_coords=[0, 0, -8],
draw_cross=False,
cmap="RdBu_r",
)
print("Description:")
pprint(results.description_)
print("References:")
pprint(results.bibtex_)
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/_utils/niimg.py:61: UserWarning: Non-finite values detected. These values will be replaced with zeros.
warn(
Description:
('An image-based meta-analysis was performed with NiMARE '
'0.2.2+0.g07ac3b6.dirty (RRID:SCR_017398; \\citealt{Salo2023}), on 21 beta '
'and variance images using the Hedges method \\citep{hedges2014statistical}, '
'in which tau-squared is estimated on a voxel-wise basis.')
References:
('@article{Salo2023,\n'
' doi = {10.52294/001c.87681},\n'
' url = {https://doi.org/10.52294/001c.87681},\n'
' year = {2023},\n'
' volume = {3},\n'
' pages = {1 - 32},\n'
' author = {Taylor Salo and Tal Yarkoni and Thomas E. Nichols and '
'Jean-Baptiste Poline and Murat Bilgel and Katherine L. Bottenhorn and Dorota '
'Jarecka and James D. Kent and Adam Kimbler and Dylan M. Nielson and Kendra '
'M. Oudyk and Julio A. Peraza and Alexandre Pérez and Puck C. Reeders and '
'Julio A. Yanes and Angela R. Laird},\n'
' title = {NiMARE: Neuroimaging Meta-Analysis Research Environment},\n'
' journal = {Aperture Neuro}\n'
'}\n'
'@book{hedges2014statistical,\n'
' title={Statistical methods for meta-analysis},\n'
' author={Hedges, Larry V and Olkin, Ingram},\n'
' year={2014},\n'
' publisher={Academic press}\n'
'}')
Total running time of the script: (1 minutes 35.328 seconds)