Note
Go to the end to download the full example code
Two-sample ALE meta-analysis
Meta-analytic projects often involve a number of common steps comparing two or more samples.
In this example, we replicate the ALE-based analyses from Enge et al.[1].
A common project workflow with two meta-analytic samples involves the following:
Run a within-sample meta-analysis of the first sample.
Characterize/summarize the results of the first meta-analysis.
Run a within-sample meta-analysis of the second sample.
Characterize/summarize the results of the second meta-analysis.
Compare the two samples with a subtraction analysis.
Compare the two within-sample meta-analyses with a conjunction analysis.
import os
from pathlib import Path
import matplotlib.pyplot as plt
from nilearn.plotting import plot_stat_map
Load Sleuth text files into Datasets
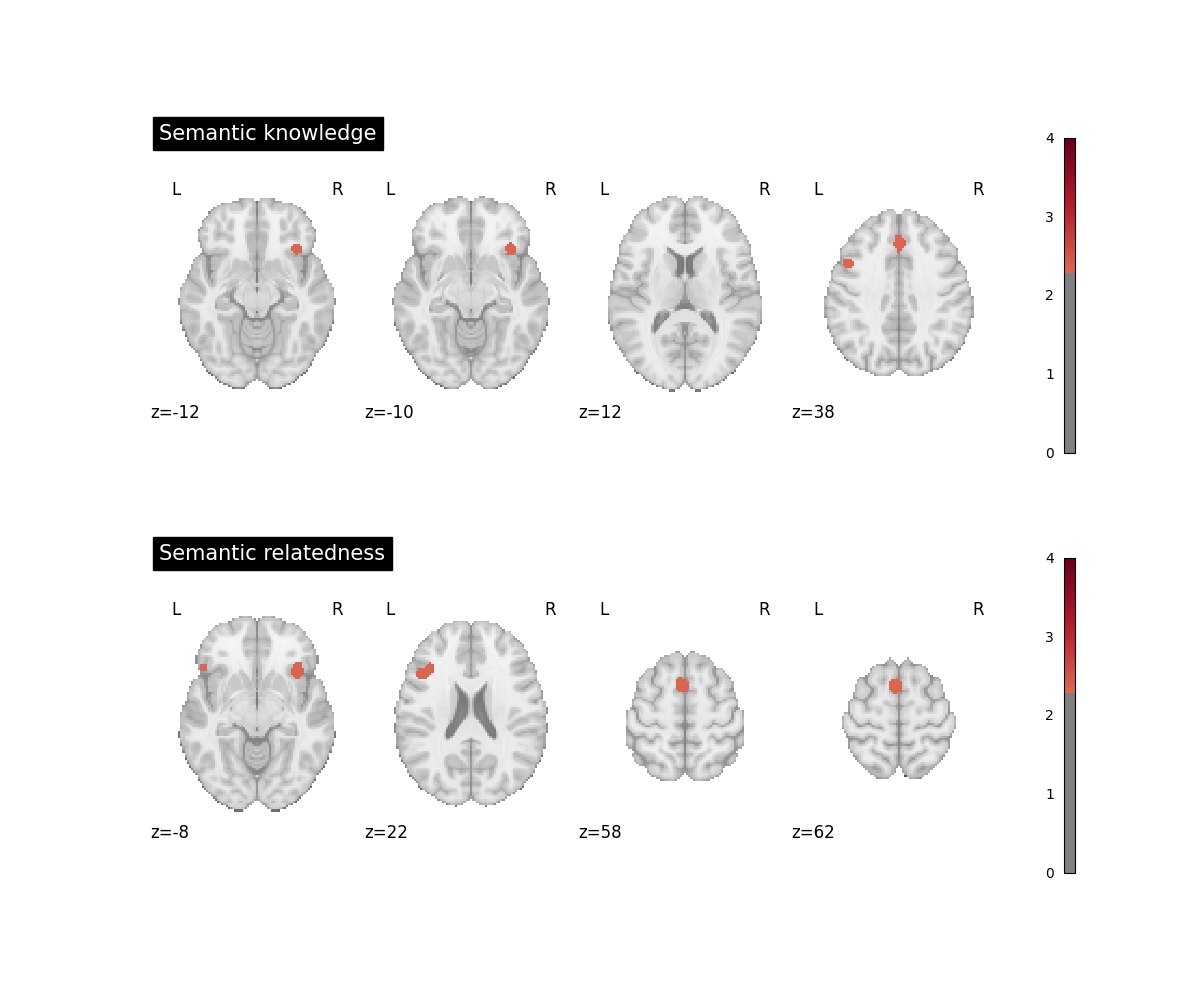
The data for this example are a subset of studies from a meta-analysis on semantic cognition in children [1]. A first group of studies probed children’s semantic world knowledge (e.g., correctly naming an object after hearing its auditory description) while a second group of studies asked children to decide if two (or more) words were semantically related to one another or not.
from nimare.io import convert_sleuth_to_dataset
from nimare.utils import get_resource_path
knowledge_file = os.path.join(get_resource_path(), "semantic_knowledge_children.txt")
related_file = os.path.join(get_resource_path(), "semantic_relatedness_children.txt")
knowledge_dset = convert_sleuth_to_dataset(knowledge_file)
related_dset = convert_sleuth_to_dataset(related_file)
Individual group ALEs
Computing separate ALE analyses for each group is not strictly necessary for performing the subtraction analysis but will help the experimenter to appreciate the similarities and differences between the groups.
from nimare.correct import FWECorrector
from nimare.meta.cbma import ALE
ale = ALE(null_method="approximate")
knowledge_results = ale.fit(knowledge_dset)
related_results = ale.fit(related_dset)
corr = FWECorrector(method="montecarlo", voxel_thresh=0.001, n_iters=100, n_cores=2)
knowledge_corrected_results = corr.transform(knowledge_results)
related_corrected_results = corr.transform(related_results)
fig, axes = plt.subplots(figsize=(12, 10), nrows=2)
knowledge_img = knowledge_corrected_results.get_map(
"z_desc-size_level-cluster_corr-FWE_method-montecarlo"
)
plot_stat_map(
knowledge_img,
cut_coords=4,
display_mode="z",
title="Semantic knowledge",
threshold=2.326, # cluster-level p < .01, one-tailed
cmap="RdBu_r",
vmax=4,
axes=axes[0],
figure=fig,
)
related_img = related_corrected_results.get_map(
"z_desc-size_level-cluster_corr-FWE_method-montecarlo"
)
plot_stat_map(
related_img,
cut_coords=4,
display_mode="z",
title="Semantic relatedness",
threshold=2.326, # cluster-level p < .01, one-tailed
cmap="RdBu_r",
vmax=4,
axes=axes[1],
figure=fig,
)
fig.show()
0%| | 0/100 [00:00<?, ?it/s]
1%| | 1/100 [00:00<01:35, 1.03it/s]
3%|▎ | 3/100 [00:02<01:02, 1.55it/s]
4%|▍ | 4/100 [00:02<00:47, 2.01it/s]
5%|▌ | 5/100 [00:02<00:54, 1.74it/s]
6%|▌ | 6/100 [00:03<00:49, 1.91it/s]
7%|▋ | 7/100 [00:04<00:52, 1.76it/s]
8%|▊ | 8/100 [00:04<00:46, 1.99it/s]
9%|▉ | 9/100 [00:05<00:48, 1.89it/s]
10%|█ | 10/100 [00:05<00:48, 1.86it/s]
11%|█ | 11/100 [00:06<00:48, 1.85it/s]
12%|█▏ | 12/100 [00:06<00:44, 1.99it/s]
13%|█▎ | 13/100 [00:07<00:44, 1.98it/s]
14%|█▍ | 14/100 [00:07<00:46, 1.86it/s]
15%|█▌ | 15/100 [00:08<00:43, 1.94it/s]
16%|█▌ | 16/100 [00:08<00:42, 1.97it/s]
17%|█▋ | 17/100 [00:09<00:40, 2.04it/s]
18%|█▊ | 18/100 [00:09<00:45, 1.82it/s]
19%|█▉ | 19/100 [00:10<00:41, 1.96it/s]
20%|██ | 20/100 [00:10<00:41, 1.91it/s]
21%|██ | 21/100 [00:11<00:37, 2.10it/s]
22%|██▏ | 22/100 [00:11<00:44, 1.75it/s]
23%|██▎ | 23/100 [00:12<00:37, 2.04it/s]
24%|██▍ | 24/100 [00:12<00:41, 1.82it/s]
25%|██▌ | 25/100 [00:13<00:35, 2.14it/s]
26%|██▌ | 26/100 [00:14<00:43, 1.68it/s]
27%|██▋ | 27/100 [00:14<00:34, 2.10it/s]
28%|██▊ | 28/100 [00:15<00:45, 1.58it/s]
30%|███ | 30/100 [00:16<00:41, 1.68it/s]
32%|███▏ | 32/100 [00:17<00:37, 1.83it/s]
33%|███▎ | 33/100 [00:17<00:29, 2.23it/s]
34%|███▍ | 34/100 [00:18<00:38, 1.71it/s]
35%|███▌ | 35/100 [00:18<00:31, 2.08it/s]
36%|███▌ | 36/100 [00:19<00:36, 1.78it/s]
37%|███▋ | 37/100 [00:19<00:31, 1.98it/s]
38%|███▊ | 38/100 [00:20<00:35, 1.74it/s]
39%|███▉ | 39/100 [00:20<00:29, 2.07it/s]
40%|████ | 40/100 [00:21<00:32, 1.83it/s]
41%|████ | 41/100 [00:21<00:30, 1.93it/s]
42%|████▏ | 42/100 [00:22<00:32, 1.79it/s]
43%|████▎ | 43/100 [00:22<00:27, 2.04it/s]
44%|████▍ | 44/100 [00:23<00:29, 1.90it/s]
45%|████▌ | 45/100 [00:24<00:29, 1.87it/s]
46%|████▌ | 46/100 [00:24<00:28, 1.86it/s]
47%|████▋ | 47/100 [00:25<00:29, 1.82it/s]
48%|████▊ | 48/100 [00:25<00:25, 2.00it/s]
49%|████▉ | 49/100 [00:26<00:26, 1.90it/s]
50%|█████ | 50/100 [00:26<00:26, 1.90it/s]
51%|█████ | 51/100 [00:27<00:26, 1.82it/s]
52%|█████▏ | 52/100 [00:27<00:23, 2.01it/s]
53%|█████▎ | 53/100 [00:28<00:24, 1.89it/s]
54%|█████▍ | 54/100 [00:28<00:23, 1.94it/s]
55%|█████▌ | 55/100 [00:29<00:24, 1.80it/s]
56%|█████▌ | 56/100 [00:29<00:20, 2.11it/s]
57%|█████▋ | 57/100 [00:30<00:22, 1.91it/s]
58%|█████▊ | 58/100 [00:30<00:21, 1.97it/s]
59%|█████▉ | 59/100 [00:31<00:22, 1.81it/s]
60%|██████ | 60/100 [00:31<00:18, 2.15it/s]
61%|██████ | 61/100 [00:32<00:20, 1.88it/s]
62%|██████▏ | 62/100 [00:32<00:18, 2.03it/s]
63%|██████▎ | 63/100 [00:33<00:20, 1.79it/s]
64%|██████▍ | 64/100 [00:33<00:16, 2.15it/s]
65%|██████▌ | 65/100 [00:34<00:18, 1.86it/s]
66%|██████▌ | 66/100 [00:34<00:16, 2.07it/s]
67%|██████▋ | 67/100 [00:35<00:18, 1.75it/s]
68%|██████▊ | 68/100 [00:35<00:14, 2.26it/s]
69%|██████▉ | 69/100 [00:36<00:17, 1.78it/s]
70%|███████ | 70/100 [00:36<00:13, 2.16it/s]
71%|███████ | 71/100 [00:37<00:17, 1.68it/s]
73%|███████▎ | 73/100 [00:38<00:14, 1.87it/s]
74%|███████▍ | 74/100 [00:38<00:11, 2.29it/s]
75%|███████▌ | 75/100 [00:39<00:14, 1.73it/s]
77%|███████▋ | 77/100 [00:40<00:12, 1.89it/s]
78%|███████▊ | 78/100 [00:40<00:09, 2.30it/s]
79%|███████▉ | 79/100 [00:41<00:11, 1.75it/s]
81%|████████ | 81/100 [00:42<00:10, 1.87it/s]
82%|████████▏ | 82/100 [00:42<00:07, 2.27it/s]
83%|████████▎ | 83/100 [00:43<00:09, 1.76it/s]
85%|████████▌ | 85/100 [00:44<00:08, 1.82it/s]
86%|████████▌ | 86/100 [00:44<00:06, 2.24it/s]
87%|████████▋ | 87/100 [00:45<00:07, 1.72it/s]
89%|████████▉ | 89/100 [00:47<00:06, 1.77it/s]
91%|█████████ | 91/100 [00:48<00:04, 1.85it/s]
93%|█████████▎| 93/100 [00:49<00:03, 1.84it/s]
95%|█████████▌| 95/100 [00:50<00:02, 1.94it/s]
97%|█████████▋| 97/100 [00:51<00:01, 1.90it/s]
99%|█████████▉| 99/100 [00:52<00:00, 1.94it/s]
100%|██████████| 100/100 [00:52<00:00, 2.22it/s]
100%|██████████| 100/100 [00:52<00:00, 1.91it/s]
0%| | 0/100 [00:00<?, ?it/s]
1%| | 1/100 [00:00<01:28, 1.12it/s]
3%|▎ | 3/100 [00:01<00:57, 1.70it/s]
5%|▌ | 5/100 [00:02<00:46, 2.04it/s]
7%|▋ | 7/100 [00:03<00:45, 2.05it/s]
9%|▉ | 9/100 [00:04<00:41, 2.20it/s]
11%|█ | 11/100 [00:05<00:41, 2.16it/s]
13%|█▎ | 13/100 [00:06<00:38, 2.26it/s]
15%|█▌ | 15/100 [00:07<00:38, 2.19it/s]
16%|█▌ | 16/100 [00:07<00:32, 2.57it/s]
17%|█▋ | 17/100 [00:07<00:38, 2.18it/s]
18%|█▊ | 18/100 [00:08<00:30, 2.66it/s]
19%|█▉ | 19/100 [00:08<00:40, 2.02it/s]
20%|██ | 20/100 [00:09<00:31, 2.51it/s]
21%|██ | 21/100 [00:09<00:37, 2.08it/s]
22%|██▏ | 22/100 [00:09<00:29, 2.67it/s]
23%|██▎ | 23/100 [00:10<00:39, 1.94it/s]
24%|██▍ | 24/100 [00:10<00:31, 2.43it/s]
25%|██▌ | 25/100 [00:11<00:36, 2.08it/s]
26%|██▌ | 26/100 [00:11<00:27, 2.65it/s]
27%|██▋ | 27/100 [00:12<00:37, 1.96it/s]
28%|██▊ | 28/100 [00:12<00:30, 2.34it/s]
29%|██▉ | 29/100 [00:13<00:33, 2.12it/s]
30%|███ | 30/100 [00:13<00:27, 2.51it/s]
31%|███ | 31/100 [00:14<00:34, 1.98it/s]
32%|███▏ | 32/100 [00:14<00:29, 2.28it/s]
33%|███▎ | 33/100 [00:15<00:31, 2.12it/s]
34%|███▍ | 34/100 [00:15<00:27, 2.39it/s]
35%|███▌ | 35/100 [00:16<00:31, 2.04it/s]
36%|███▌ | 36/100 [00:16<00:27, 2.30it/s]
37%|███▋ | 37/100 [00:16<00:28, 2.22it/s]
38%|███▊ | 38/100 [00:17<00:26, 2.38it/s]
39%|███▉ | 39/100 [00:17<00:29, 2.09it/s]
40%|████ | 40/100 [00:18<00:27, 2.21it/s]
41%|████ | 41/100 [00:18<00:26, 2.24it/s]
42%|████▏ | 42/100 [00:19<00:25, 2.28it/s]
43%|████▎ | 43/100 [00:19<00:26, 2.11it/s]
44%|████▍ | 44/100 [00:20<00:26, 2.14it/s]
45%|████▌ | 45/100 [00:20<00:23, 2.31it/s]
46%|████▌ | 46/100 [00:20<00:24, 2.24it/s]
47%|████▋ | 47/100 [00:21<00:23, 2.23it/s]
48%|████▊ | 48/100 [00:21<00:24, 2.13it/s]
49%|████▉ | 49/100 [00:22<00:21, 2.40it/s]
50%|█████ | 50/100 [00:22<00:22, 2.18it/s]
51%|█████ | 51/100 [00:23<00:21, 2.24it/s]
52%|█████▏ | 52/100 [00:23<00:22, 2.09it/s]
53%|█████▎ | 53/100 [00:23<00:19, 2.41it/s]
54%|█████▍ | 54/100 [00:24<00:21, 2.15it/s]
55%|█████▌ | 55/100 [00:24<00:20, 2.23it/s]
56%|█████▌ | 56/100 [00:25<00:21, 2.05it/s]
57%|█████▋ | 57/100 [00:25<00:18, 2.35it/s]
58%|█████▊ | 58/100 [00:26<00:19, 2.21it/s]
59%|█████▉ | 59/100 [00:26<00:19, 2.14it/s]
60%|██████ | 60/100 [00:27<00:18, 2.16it/s]
61%|██████ | 61/100 [00:27<00:17, 2.28it/s]
62%|██████▏ | 62/100 [00:28<00:16, 2.25it/s]
63%|██████▎ | 63/100 [00:28<00:17, 2.12it/s]
64%|██████▍ | 64/100 [00:29<00:16, 2.18it/s]
65%|██████▌ | 65/100 [00:29<00:15, 2.28it/s]
66%|██████▌ | 66/100 [00:29<00:15, 2.27it/s]
67%|██████▋ | 67/100 [00:30<00:15, 2.13it/s]
68%|██████▊ | 68/100 [00:30<00:14, 2.18it/s]
69%|██████▉ | 69/100 [00:31<00:13, 2.31it/s]
70%|███████ | 70/100 [00:31<00:13, 2.30it/s]
71%|███████ | 71/100 [00:32<00:13, 2.15it/s]
72%|███████▏ | 72/100 [00:32<00:12, 2.18it/s]
73%|███████▎ | 73/100 [00:33<00:11, 2.29it/s]
74%|███████▍ | 74/100 [00:33<00:11, 2.27it/s]
75%|███████▌ | 75/100 [00:34<00:11, 2.11it/s]
76%|███████▌ | 76/100 [00:34<00:11, 2.16it/s]
77%|███████▋ | 77/100 [00:34<00:09, 2.31it/s]
78%|███████▊ | 78/100 [00:35<00:09, 2.26it/s]
79%|███████▉ | 79/100 [00:35<00:09, 2.10it/s]
80%|████████ | 80/100 [00:36<00:09, 2.16it/s]
81%|████████ | 81/100 [00:36<00:08, 2.31it/s]
82%|████████▏ | 82/100 [00:37<00:08, 2.24it/s]
83%|████████▎ | 83/100 [00:37<00:07, 2.23it/s]
84%|████████▍ | 84/100 [00:38<00:07, 2.15it/s]
85%|████████▌ | 85/100 [00:38<00:06, 2.40it/s]
86%|████████▌ | 86/100 [00:38<00:06, 2.21it/s]
87%|████████▋ | 87/100 [00:39<00:05, 2.20it/s]
88%|████████▊ | 88/100 [00:39<00:05, 2.08it/s]
89%|████████▉ | 89/100 [00:40<00:04, 2.35it/s]
90%|█████████ | 90/100 [00:40<00:04, 2.18it/s]
91%|█████████ | 91/100 [00:41<00:04, 2.21it/s]
92%|█████████▏| 92/100 [00:41<00:03, 2.08it/s]
93%|█████████▎| 93/100 [00:42<00:02, 2.35it/s]
94%|█████████▍| 94/100 [00:42<00:02, 2.16it/s]
95%|█████████▌| 95/100 [00:43<00:02, 2.17it/s]
96%|█████████▌| 96/100 [00:43<00:01, 2.11it/s]
97%|█████████▋| 97/100 [00:43<00:01, 2.38it/s]
98%|█████████▊| 98/100 [00:44<00:00, 2.21it/s]
99%|█████████▉| 99/100 [00:44<00:00, 2.19it/s]
100%|██████████| 100/100 [00:45<00:00, 2.27it/s]
100%|██████████| 100/100 [00:45<00:00, 2.21it/s]
Characterize the relative contributions of experiments in the ALE results
NiMARE contains two methods for this: Jackknife
and FocusCounter.
We will show both below, but for the sake of speed we will only apply one to
each subgroup meta-analysis.
from nimare.diagnostics import FocusCounter
counter = FocusCounter(
target_image="z_desc-size_level-cluster_corr-FWE_method-montecarlo",
voxel_thresh=None,
)
knowledge_diagnostic_results = counter.transform(knowledge_corrected_results)
0%| | 0/21 [00:00<?, ?it/s]
10%|▉ | 2/21 [00:00<00:01, 10.87it/s]
19%|█▉ | 4/21 [00:00<00:01, 9.69it/s]
24%|██▍ | 5/21 [00:00<00:01, 9.51it/s]
29%|██▊ | 6/21 [00:00<00:01, 9.37it/s]
33%|███▎ | 7/21 [00:00<00:01, 9.24it/s]
38%|███▊ | 8/21 [00:00<00:01, 9.19it/s]
43%|████▎ | 9/21 [00:00<00:01, 9.12it/s]
48%|████▊ | 10/21 [00:01<00:01, 9.09it/s]
52%|█████▏ | 11/21 [00:01<00:01, 9.04it/s]
57%|█████▋ | 12/21 [00:01<00:00, 9.02it/s]
62%|██████▏ | 13/21 [00:01<00:00, 9.03it/s]
67%|██████▋ | 14/21 [00:01<00:00, 8.98it/s]
71%|███████▏ | 15/21 [00:01<00:00, 9.01it/s]
76%|███████▌ | 16/21 [00:01<00:00, 8.91it/s]
81%|████████ | 17/21 [00:01<00:00, 8.99it/s]
86%|████████▌ | 18/21 [00:01<00:00, 8.98it/s]
90%|█████████ | 19/21 [00:02<00:00, 8.96it/s]
95%|█████████▌| 20/21 [00:02<00:00, 8.83it/s]
100%|██████████| 21/21 [00:02<00:00, 8.90it/s]
100%|██████████| 21/21 [00:02<00:00, 9.12it/s]
Clusters table.
knowledge_clusters_table = knowledge_diagnostic_results.tables[
"z_desc-size_level-cluster_corr-FWE_method-montecarlo_tab-clust"
]
knowledge_clusters_table.head(10)
Contribution table. Here PostiveTail refers to clusters with positive statistics.
knowledge_count_table = knowledge_diagnostic_results.tables[
"z_desc-size_level-cluster_corr-FWE_method-montecarlo_diag-FocusCounter"
"_tab-counts_tail-positive"
]
knowledge_count_table.head(10)
from nimare.diagnostics import Jackknife
jackknife = Jackknife(
target_image="z_desc-size_level-cluster_corr-FWE_method-montecarlo",
voxel_thresh=None,
)
related_diagnostic_results = jackknife.transform(related_corrected_results)
related_jackknife_table = related_diagnostic_results.tables[
"z_desc-size_level-cluster_corr-FWE_method-montecarlo_diag-Jackknife_tab-counts_tail-positive"
]
related_jackknife_table.head(10)
0%| | 0/16 [00:00<?, ?it/s]
6%|▋ | 1/16 [00:02<00:30, 2.01s/it]
12%|█▎ | 2/16 [00:04<00:28, 2.02s/it]
19%|█▉ | 3/16 [00:05<00:25, 1.99s/it]
25%|██▌ | 4/16 [00:08<00:24, 2.00s/it]
31%|███▏ | 5/16 [00:10<00:22, 2.02s/it]
38%|███▊ | 6/16 [00:12<00:20, 2.06s/it]
44%|████▍ | 7/16 [00:14<00:18, 2.08s/it]
50%|█████ | 8/16 [00:16<00:16, 2.07s/it]
56%|█████▋ | 9/16 [00:18<00:14, 2.06s/it]
62%|██████▎ | 10/16 [00:20<00:12, 2.06s/it]
69%|██████▉ | 11/16 [00:22<00:10, 2.04s/it]
75%|███████▌ | 12/16 [00:24<00:08, 2.03s/it]
81%|████████▏ | 13/16 [00:26<00:06, 2.01s/it]
88%|████████▊ | 14/16 [00:28<00:03, 2.00s/it]
94%|█████████▍| 15/16 [00:30<00:01, 1.99s/it]
100%|██████████| 16/16 [00:32<00:00, 1.98s/it]
100%|██████████| 16/16 [00:32<00:00, 2.02s/it]
Subtraction analysis
Typically, one would use at least 5000 iterations for a subtraction analysis. However, we have reduced this to 100 iterations for this example. Similarly here we use a voxel-level z-threshold of 0.01, but in practice one would use a more stringent threshold (e.g., 1.65).
from nimare.meta.cbma import ALESubtraction
from nimare.reports.base import run_reports
from nimare.workflows import PairwiseCBMAWorkflow
workflow = PairwiseCBMAWorkflow(
estimator=ALESubtraction(n_iters=10, n_cores=1),
corrector="fdr",
diagnostics=FocusCounter(voxel_thresh=0.01, display_second_group=True),
)
res_sub = workflow.fit(knowledge_dset, related_dset)
0%| | 0/10 [00:00<?, ?it/s]
10%|█ | 1/10 [00:00<00:04, 1.82it/s]
20%|██ | 2/10 [00:01<00:04, 1.82it/s]
30%|███ | 3/10 [00:01<00:03, 1.81it/s]
40%|████ | 4/10 [00:02<00:03, 1.82it/s]
50%|█████ | 5/10 [00:02<00:02, 1.82it/s]
60%|██████ | 6/10 [00:03<00:02, 1.84it/s]
70%|███████ | 7/10 [00:03<00:01, 1.83it/s]
80%|████████ | 8/10 [00:04<00:01, 1.83it/s]
90%|█████████ | 9/10 [00:04<00:00, 1.82it/s]
100%|██████████| 10/10 [00:05<00:00, 1.82it/s]
100%|██████████| 10/10 [00:05<00:00, 1.82it/s]
0%| | 0/228483 [00:00<?, ?it/s]
0%| | 2/228483 [00:00<14:00, 271.79it/s]
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/reporting/_get_clusters_table.py:104: UserWarning: Attention: At least one of the (sub)peaks falls outside of the cluster body. Identifying the nearest in-cluster voxel.
warnings.warn(
0%| | 0/21 [00:00<?, ?it/s]
5%|▍ | 1/21 [00:00<00:02, 7.26it/s]
10%|▉ | 2/21 [00:00<00:02, 7.25it/s]
14%|█▍ | 3/21 [00:00<00:02, 7.39it/s]
19%|█▉ | 4/21 [00:00<00:02, 7.11it/s]
24%|██▍ | 5/21 [00:00<00:02, 7.24it/s]
29%|██▊ | 6/21 [00:00<00:02, 7.25it/s]
33%|███▎ | 7/21 [00:00<00:01, 7.16it/s]
38%|███▊ | 8/21 [00:01<00:01, 7.08it/s]
43%|████▎ | 9/21 [00:01<00:01, 7.26it/s]
48%|████▊ | 10/21 [00:01<00:01, 7.13it/s]
52%|█████▏ | 11/21 [00:01<00:01, 7.00it/s]
57%|█████▋ | 12/21 [00:01<00:01, 7.13it/s]
62%|██████▏ | 13/21 [00:01<00:01, 7.00it/s]
67%|██████▋ | 14/21 [00:01<00:00, 7.04it/s]
71%|███████▏ | 15/21 [00:02<00:00, 7.05it/s]
76%|███████▌ | 16/21 [00:02<00:00, 7.04it/s]
81%|████████ | 17/21 [00:02<00:00, 7.30it/s]
86%|████████▌ | 18/21 [00:02<00:00, 7.38it/s]
90%|█████████ | 19/21 [00:02<00:00, 7.23it/s]
95%|█████████▌| 20/21 [00:02<00:00, 7.34it/s]
100%|██████████| 21/21 [00:02<00:00, 7.21it/s]
100%|██████████| 21/21 [00:02<00:00, 7.17it/s]
0%| | 0/16 [00:00<?, ?it/s]
6%|▋ | 1/16 [00:00<00:02, 7.09it/s]
12%|█▎ | 2/16 [00:00<00:01, 7.20it/s]
19%|█▉ | 3/16 [00:00<00:01, 7.02it/s]
25%|██▌ | 4/16 [00:00<00:01, 7.04it/s]
31%|███▏ | 5/16 [00:00<00:01, 6.98it/s]
38%|███▊ | 6/16 [00:00<00:01, 6.93it/s]
44%|████▍ | 7/16 [00:01<00:01, 6.90it/s]
50%|█████ | 8/16 [00:01<00:01, 6.96it/s]
56%|█████▋ | 9/16 [00:01<00:00, 7.02it/s]
62%|██████▎ | 10/16 [00:01<00:00, 6.91it/s]
69%|██████▉ | 11/16 [00:01<00:00, 6.89it/s]
75%|███████▌ | 12/16 [00:01<00:00, 6.93it/s]
81%|████████▏ | 13/16 [00:01<00:00, 6.96it/s]
88%|████████▊ | 14/16 [00:02<00:00, 6.99it/s]
94%|█████████▍| 15/16 [00:02<00:00, 6.91it/s]
100%|██████████| 16/16 [00:02<00:00, 6.88it/s]
100%|██████████| 16/16 [00:02<00:00, 6.95it/s]
Report
Finally, a NiMARE report is generated from the MetaResult. root_dir = Path(os.getcwd()).parents[1] / “docs” / “_build” Use the previous root to run the documentation locally.
root_dir = Path(os.getcwd()).parents[1] / "_readthedocs"
html_dir = root_dir / "html" / "auto_examples" / "02_meta-analyses" / "08_subtraction"
html_dir.mkdir(parents=True, exist_ok=True)
run_reports(res_sub, html_dir)
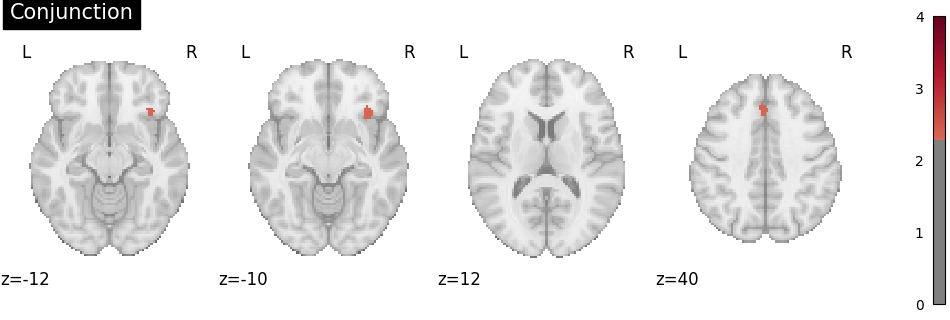
Conjunction analysis
To determine the overlap of the meta-analytic results, a conjunction image can be computed by (a) identifying voxels that were statistically significant in both individual group maps and (b) selecting, for each of these voxels, the smaller of the two group-specific z values Nichols et al.[2].
from nimare.workflows.misc import conjunction_analysis
img_conj = conjunction_analysis([knowledge_img, related_img])
plot_stat_map(
img_conj,
cut_coords=4,
display_mode="z",
title="Conjunction",
threshold=2.326, # cluster-level p < .01, one-tailed
cmap="RdBu_r",
vmax=4,
)
<nilearn.plotting.displays._slicers.ZSlicer object at 0x7ffa48824100>
References
Total running time of the script: (3 minutes 55.927 seconds)