Note
Go to the end to download the full example code
GCLDA topic modeling
Train a generalized correspondence latent Dirichlet allocation model using abstracts.
Warning
The model in this example is trained using (1) a very small, nonrepresentative dataset and (2) very few iterations. As such, it will not provide useful results. If you are interested in using GCLDA, we recommend using a large dataset like Neurosynth, and training with at least 10k iterations.
import os
import nibabel as nib
import numpy as np
from nilearn import image, masking, plotting
from nimare import annotate, decode
from nimare.dataset import Dataset
from nimare.utils import get_resource_path
Load dataset with abstracts
We’ll load a small dataset composed only of studies in Neurosynth with Angela Laird as a coauthor, for the sake of speed.
dset = Dataset(os.path.join(get_resource_path(), "neurosynth_laird_studies.json"))
dset.texts.head(2)
Generate term counts
GCLDA uses raw word counts instead of the tf-idf values generated by Neurosynth.
counts_df = annotate.text.generate_counts(
dset.texts,
text_column="abstract",
tfidf=False,
max_df=0.99,
min_df=0.01,
)
counts_df.head(5)
Run model
Five iterations will take ~10 minutes with the full Neurosynth dataset. It’s much faster with this reduced example dataset. Note that we’re using only 10 topics here. This is because there are only 13 studies in the dataset. If the number of topics is higher than the number of studies in the dataset, errors can occur during training.
model = annotate.gclda.GCLDAModel(
counts_df,
dset.coordinates,
mask=dset.masker.mask_img,
n_topics=10,
n_regions=4,
symmetric=True,
)
model.fit(n_iters=100, loglikely_freq=20)
model.save("gclda_model.pkl.gz")
# Let's remove the model now that you know how to generate it.
os.remove("gclda_model.pkl.gz")
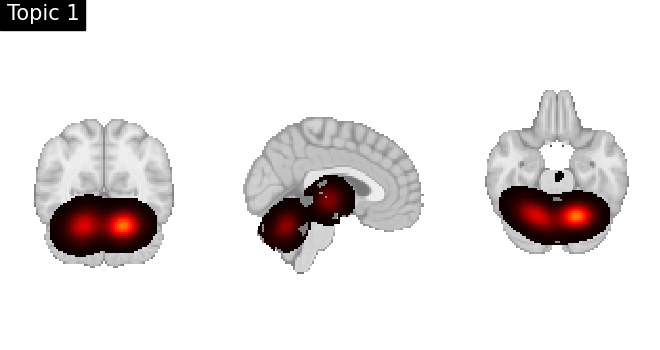
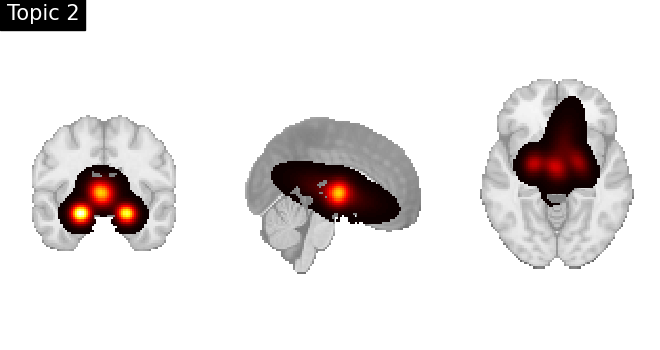
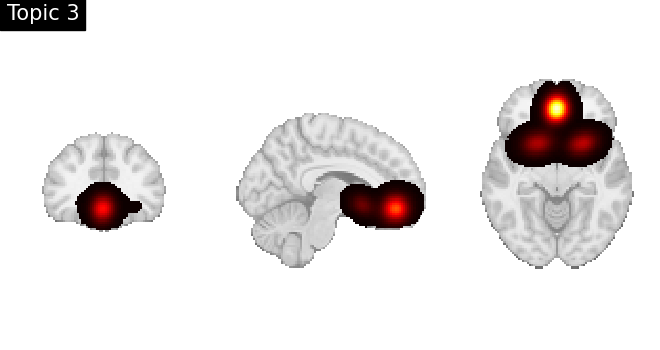
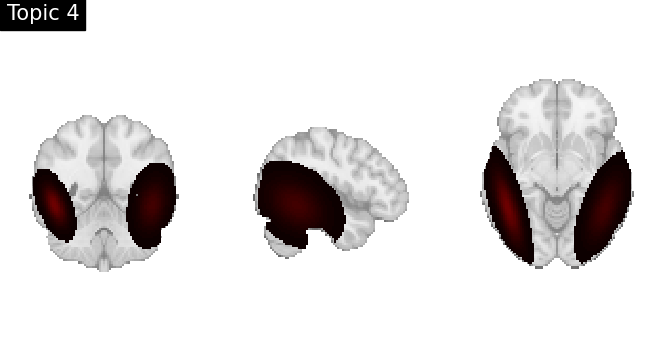
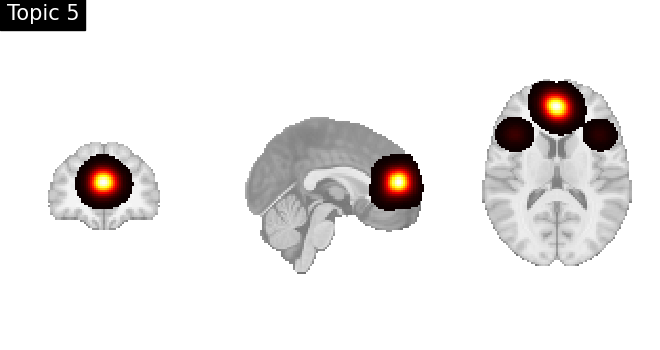
Look at topics
topic_img_4d = masking.unmask(model.p_voxel_g_topic_.T, model.mask)
for i_topic in range(5):
topic_img_3d = image.index_img(topic_img_4d, i_topic)
plotting.plot_stat_map(
topic_img_3d,
draw_cross=False,
colorbar=False,
annotate=False,
title=f"Topic {i_topic + 1}",
)
Generate a pseudo-statistic image from text
text = "dorsal anterior cingulate cortex"
encoded_img, _ = decode.encode.gclda_encode(model, text)
plotting.plot_stat_map(encoded_img, draw_cross=False)
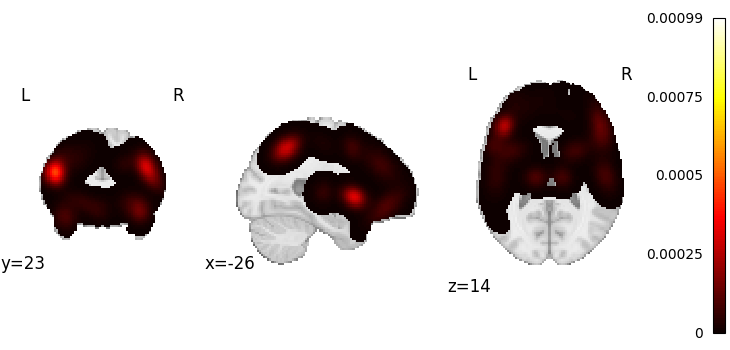
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7ffa4920f2e0>
Decode an unthresholded statistical map
For the sake of simplicity, we will use the pseudo-statistic map generated in the previous step.
# Run the decoder
decoded_df, _ = decode.continuous.gclda_decode_map(model, encoded_img)
decoded_df.sort_values(by="Weight", ascending=False).head(10)
Decode an ROI image
First we’ll make an ROI
arr = np.zeros(dset.masker.mask_img.shape, np.int32)
arr[65:75, 50:60, 50:60] = 1
mask_img = nib.Nifti1Image(arr, dset.masker.mask_img.affine)
plotting.plot_roi(mask_img, draw_cross=False)
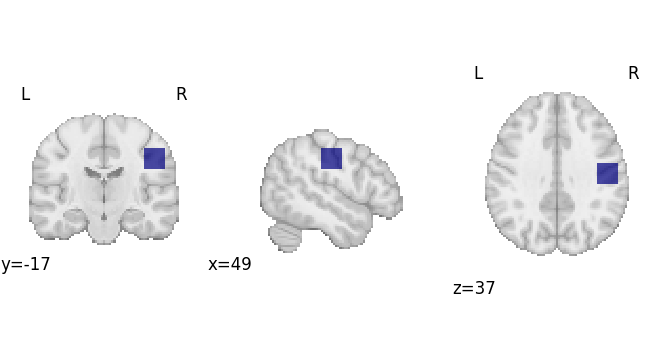
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7ffa48ee7fd0>
Run the decoder
decoded_df, _ = decode.discrete.gclda_decode_roi(model, mask_img)
decoded_df.sort_values(by="Weight", ascending=False).head(10)
Total running time of the script: (0 minutes 38.878 seconds)