Note
Go to the end to download the full example code
Run an image-based meta-analysis (IBMA) workflow
NiMARE provides a plethora of tools for performing meta-analyses on neuroimaging data. Sometimes it’s difficult to know where to start, especially if you’re new to meta-analysis. This tutorial will walk you through using a IBMA workflow function which puts together the fundamental steps of a IBMA meta-analysis.
import os
from pathlib import Path
import matplotlib.pyplot as plt
from nilearn.plotting import plot_stat_map
from nimare.extract import download_nidm_pain
Download data
Load Dataset
from nimare.dataset import Dataset
from nimare.utils import get_resource_path
dset_file = os.path.join(get_resource_path(), "nidm_pain_dset.json")
dset = Dataset(dset_file)
dset.update_path(dset_dir)
Run IBMA Workflow
The fit method of a IBMA workflow class runs the following steps:
Runs a meta-analysis using the specified method (default: Stouffers)
Applies a corrector to the meta-analysis results (default: FDRCorrector, indep)
Generates cluster tables and runs diagnostics on the corrected results (default: Jackknife)
All in one call!
from nimare.workflows.ibma import IBMAWorkflow
workflow = IBMAWorkflow()
result = workflow.fit(dset)
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning:
imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning:
imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning:
imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning:
imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning:
imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning:
imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning:
imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning:
imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning:
imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning:
imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/_utils/niimg.py:61: UserWarning:
Non-finite values detected. These values will be replaced with zeros.
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/reporting/_get_clusters_table.py:339: UserWarning:
Attention: No clusters with stat lower than -1.65
0%| | 0/21 [00:00<?, ?it/s]
5%|▍ | 1/21 [00:04<01:37, 4.86s/it]
10%|▉ | 2/21 [00:09<01:32, 4.89s/it]
14%|█▍ | 3/21 [00:14<01:27, 4.86s/it]
19%|█▉ | 4/21 [00:19<01:22, 4.85s/it]
24%|██▍ | 5/21 [00:24<01:17, 4.83s/it]
29%|██▊ | 6/21 [00:28<01:12, 4.81s/it]
33%|███▎ | 7/21 [00:33<01:07, 4.80s/it]
38%|███▊ | 8/21 [00:38<01:02, 4.81s/it]
43%|████▎ | 9/21 [00:43<00:57, 4.82s/it]
48%|████▊ | 10/21 [00:48<00:53, 4.83s/it]
52%|█████▏ | 11/21 [00:53<00:47, 4.80s/it]
57%|█████▋ | 12/21 [00:57<00:43, 4.79s/it]
62%|██████▏ | 13/21 [01:02<00:38, 4.80s/it]
67%|██████▋ | 14/21 [01:07<00:33, 4.77s/it]
71%|███████▏ | 15/21 [01:12<00:28, 4.76s/it]
76%|███████▌ | 16/21 [01:17<00:24, 4.81s/it]
81%|████████ | 17/21 [01:21<00:19, 4.82s/it]
86%|████████▌ | 18/21 [01:26<00:14, 4.81s/it]
90%|█████████ | 19/21 [01:31<00:09, 4.80s/it]
95%|█████████▌| 20/21 [01:36<00:04, 4.79s/it]
100%|██████████| 21/21 [01:40<00:00, 4.78s/it]
100%|██████████| 21/21 [01:40<00:00, 4.81s/it]
Plot Results
The fit method of the IBMA workflow class returns a MetaResult object,
where you can access the corrected results of the meta-analysis and diagnostics tables.
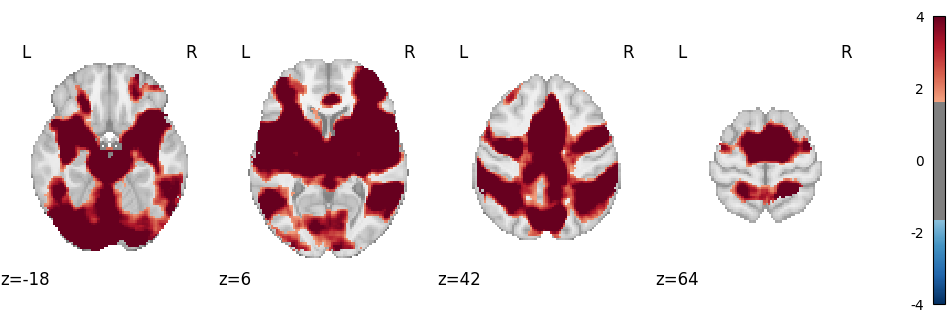
Corrected map:
img = result.get_map("z_corr-FDR_method-indep")
plot_stat_map(
img,
cut_coords=4,
display_mode="z",
threshold=1.65, # voxel_thresh p < .05, one-tailed
cmap="RdBu_r",
vmax=4,
)
plt.show()
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/_utils/niimg.py:61: UserWarning:
Non-finite values detected. These values will be replaced with zeros.
Clusters table
result.tables["z_corr-FDR_method-indep_tab-clust"]
Contribution table
result.tables["z_corr-FDR_method-indep_diag-Jackknife_tab-counts_tail-positive"]
Report
Finally, a NiMARE report is generated from the MetaResult.
from nimare.reports.base import run_reports
# root_dir = Path(os.getcwd()).parents[1] / "docs" / "_build"
# Use the previous root to run the documentation locally.
root_dir = Path(os.getcwd()).parents[1] / "_readthedocs"
html_dir = root_dir / "html" / "auto_examples" / "02_meta-analyses" / "12_plot_ibma_workflow"
html_dir.mkdir(parents=True, exist_ok=True)
run_reports(result, html_dir)
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/_utils/niimg.py:61: UserWarning:
Non-finite values detected. These values will be replaced with zeros.
/home/docs/checkouts/readthedocs.org/user_builds/nimare/envs/stable/lib/python3.9/site-packages/nilearn/_utils/niimg.py:61: UserWarning:
Non-finite values detected. These values will be replaced with zeros.
Total running time of the script: (2 minutes 58.126 seconds)